Liquid biopsy holds many advantages over solid tumor testing of cancer patients. It is less invasive for the patient, avoiding the need for surgical biopsy, and has improved levels of sensitivity to detect low frequency somatic driver mutations.
The industry is pushing for liquid biopsy to become the go-to method for the collection of clinical DNA samples for oncology genotyping. This could be at the point of diagnosis - for routine monitoring during treatment to detect the appearance of resistance mutations, which may indicate a change of therapy is required. Alternatively, liquid biopsy could be used for future preventative cancer screening of the general population before disease symptoms appear - to facilitate earlier diagnosis and better treatment outcomes.
Using cfDNA for patient diagnosis via liquid biopsy has its challenges
As with any new sequencing technology, common technical hurdles include:
- Sample handling Liquid biopsy workflows involve additional steps for sample handling. For example, clinical labs that have been more familiar with handling robust FFPE blocks for many decades are now faced with processing blood samples with shorter shelf lives, and that require multiple extraction steps such as plasma from whole blood and cfDNA from plasma. All of these extraction steps need to be properly validated to ensure they do not introduce errors in the final results.
- Reliability of results Liquid biopsy assays are required to operate with much more sensitive limits of detection than previous FFPE-based sequencing assays. As such, the technology needs to be rigorously tested to ensure it can accurately call variants down to between 5 - 0.1% allele frequency, without calling false positives.
- Sample Variability Human plasma can naturally display high lot-to-lot variability, making it difficult to control and implement a consistent protocol. In addition, high sample variation can also be introduced by inconsistencies in the clinical blood draw and immediate blood storage process, which can vary between phlebotomists and hospitals. Controlling for this variation and introducing a consistent protocol is essential for the success of wide-scale liquid biopsy adoption.
The two big challenges of liquid biopsy
- Limit of detection (LOD) and false-positive error rates
- The reliable limit of detection for this cfDNA assay is 1% AF
- They are calling a false positive for NRAS A59T
- The variability and instability of human plasma
- 400 ng of cfDNA was spiked into 1 ml of either human or synthetic plasma, and stored at -80˚C
- cfDNA was extracted by using Circulating Nucleic Acid kit (Qiagen), extraction efficiency was measured with Qubit BR Reagents (Molecular Probes)
- Total AKT1 gene copies were quantified by ddPCR (Biorad)
One of the key challenges of using cfDNA to detect cancers early, is the extremely low quantities of cfDNA in patient blood. This means when sequencing, laboratories are working at allelic frequencies much lower than they have ever had to before.
So, how can we be confident in our lower limit of detection and ensure we’re not seeing false positives appear in results?
Using a reference standard with a range of precisely defined allelic frequencies can help determine a true limit of detection and reduce the risk of false positives.
| Gene | Variant | Allelic Frequency | |||
|---|---|---|---|---|---|
| 5% | 1% | 0.1% | 0% (WT) | ||
| EGFR | L858R | 5.0 | 1.0 | ND | ND |
| EGFR | ΔE746-A750 | 4.9 | 0.9 | ND | ND |
| EGFR | T790M | 4.9 | 1.1 | ND | ND |
| EGFR | V769-D770ins | 5.0 | 1.0 | ND | ND |
| KRAS | G12D | 5.1 | 1.0 | ND | ND |
| NRAS | Q61K | 4.9 | 0.9 | ND | ND |
| NRAS | A59T | 5.2 | 1.1 | 0.7 | 0.7 |
| PIKC3A | E545K | 5.0 | 1.0 | ND | ND |
In this example data set, the reference standard informs the user that:
When a reference standard is run before a patient sample, you can be sure of your limit of detection for your assay. It also allows pipeline optimization – you can re-calibrate and amend your workflow to counter any false results. This gives you confidence in your results when handling real patient samples.
There are a few key challenges that come with using human plasma as a control for your cfDNA assay. Yes, human plasma matches your patient sample behaviors, but this does not always outweigh the challenges that come with using it as a reliable control for diagnosis.
| Real (Human) Plasma | Horizon's Synthetic Plasma |
|---|---|
| Variable quantity and concentrations | Defined volume and concentrations |
| Lot-to-lot variability | Lot-to-lot stability |
| Irregular supply | Reliable supply |
| Contamination with other analytes / genomic DNA | No interfering analytes / genomic DNA |
| cfDNA degradation: time-limited storage | Long-term stability of cfDNA: over 24 months |
How does this impact performance?
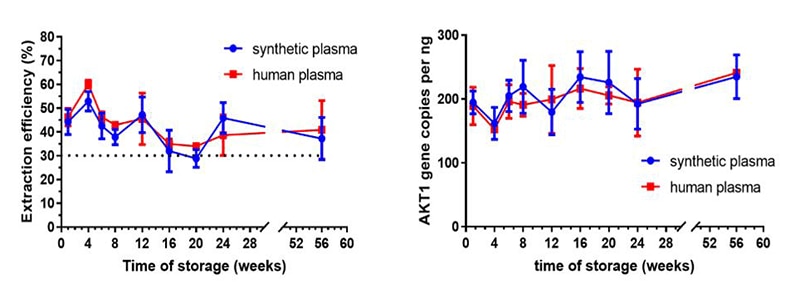
In this experiment the stability of our cfDNA spiked into either human or synthetic plasma was investigated over a 60-week period. Results show that the amount of cfDNA that could be extracted (~40% average extraction efficiency) and the gene copy number of ALK1 as determined by ddPCR, were similar in both human and synthetic plasma after 60 weeks of storage at -80°C. This demonstrates that our synthetic plasma behaves similarly and closely mimics real human plasma to act as a suitable cfDNA reference standard matrix material.
cfDNA Recovery Gene Copy Number Detection
Taking control of your workflow
Having well characterized cell-line derived reference standards that closely mimic real patient samples with clinically relevant variants defined by a gold standard mechanism like ddPCR allows a new liquid biopsy assay to be properly validated.
Users can:
- Check that their workflows accurately detect all of the variants in the control material at the correct allele frequencies without calling false positives
- Validate and control for the introduction of errors during the DNA extraction procedure
- Ensure that the design of their liquid biopsy sequencing assay is functioning effectively with no amplicon drop out (liquid biopsy assays need to sequence from smaller fragments of DNA than was previously required in fresh tissue or FFPE assays)
Horizon has developed a range of cell line-derived cfDNA reference standards to help develop, optimize, monitor and control the accuracy of new patient tests. These materials contain a range of actionable variants in key cancer genes at well characterized allele frequencies as determined by ddPCR, located within genomic DNA with an average fragment size of 160bp.
Our cfDNA material in synthetic plasma helps to monitor the entire liquid biopsy workflow, from DNA extraction to interpretation of results, giving labs confidence in the accuracy of their test.
What's next?
The future of testing, monitoring and treating cancer patients by liquid biopsy is hugely exciting. It holds promise to make genetic more accessible from a simple blood draw, and encourages more frequent testing in all aspects of cancer management including pre-disease preventative monitoring, diagnosis, treatment, tumor evolution, resistance management and long-term remission surveillance and check-up.
