Simplify CRISPR editing in all cell types, including primary cells!
The Edit-R gene editing platform enables researchers to make precise edits that result in functional gene knockout for the systematic interrogation of mammalian genome function. Horizon now offers three unique synthetic sgRNA designs per human gene, providing multiple data points for increased confidence in phenotypic results
Expanding the Edit-R product line to include predesigned synthetic sgRNAs as individual reagents and screening libraries allows researchers to:
- Streamline the CRISPR workflow – Synthetic sgRNAs improve ease of use on a trusted and long-standing gene editing platform for researchers who want a single reagent as opposed to the two-part crRNA:tracrRNA format.
- Expand possible CRISPR applications – Synthetic sgRNA is especially amenable to complex cell systems such as primary T cells, iPSCs, and other highly valuable, informative, and clinically relevant backgrounds.
- Reduce innate immune response – Synthetic guide RNAs can eliminate unwanted cellular responses caused by enzymatically derived guides.
- Guarantee CRISPR editing – Designed using our proprietary Edit-R guide RNA algorithm, all Edit-R guides are guaranteed to provide successful editing, and algorithm-optimized to maximize functional protein knockout and minimize off-target editing.
Predesigned synthetic sgRNA achieves highly effective and specific editing for robust functional gene knockout
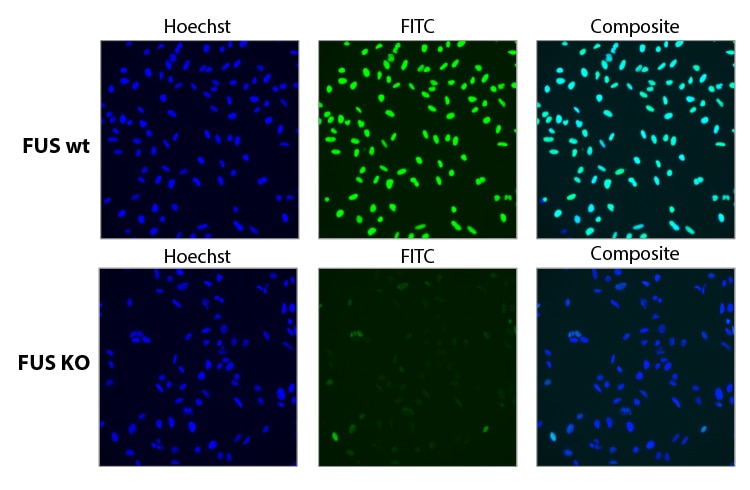
Functional knockout of FUS was assessed in U2OS cells constitutively expressing Cas9 under the CAG promoter. Cells were transfected with a 25 nM sgRNA pool containing three different Edit-R predesigned sgRNAs targeting FUS using 0.04 µL DharmaFECT 4. 72 hours post transfection the cells were split, and at 96 hours post transfection, cells were fixed and stained with a primary antibody targeting FUS, and an Alexa Fluor 488 conjugated fluorescent secondary antibody. Hoechst stain was used to identify nuclei.
Edit-R guide RNA stability modifications increase editing efficiency
All Edit-R guide RNA reagents are chemically modified to increase nuclease resistance and ensure stability throughout the rigors of delivery and the intracellular environment.
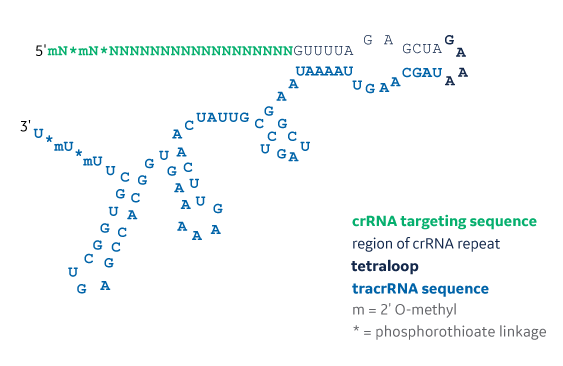
The structure of an Edit-R synthetic single guide RNA showing the 2xMS nuclease stability modifications on both 5’ and 3’ ends of the molecule. The structure contains a 20 nt targeting sequence (shown in green as poly Ns), 12 nt crRNA repeat sequence (shown in light gray), 4 nt tetraloop sequence (shown in bold black), and a 64 nt tracrRNA sequence (shown in blue). MS = 2’O-methyl nucleotides and phosphorothioate linkages (MS).
Edit-R synthetic sgRNAs arrive ready-to-use and enable rapid loss-of-function studies from individual genes and scaling up to the entire genome. Available as
- Individual sgRNA reagents
- Pools of 3 sgRNA per gene in libraries – Conveniently arrayed in 96- and 384-well plates as gene family collections, custom Cherry-pick libraries, or the entire human genome.
Search for your gene of interest or learn more about our new arrayed Edit-R sgRNA collections!
Additional Resources
- Optimized tools for high-confidence genome engineering
- Customize and order plates of predesigned crRNA for knockout studies for your targets of interest
CRISPR-Cas9 Screening libraries
- Arrayed or pooled sgRNA for high-throughput gene editing studies
