Redundancy is an inherent property of biological networks, and enables living systems to survive in response to genetic and environmental perturbations. Synthetic lethality occurs when the simultaneous disruption of two non-essential genes prohibits growth or results in death; this occurs even though the loss of either gene by itself may not cause a change in viability. The study of synthetic lethality plays a key role in identifying functional associations between genes. Synthetic lethality also occurs between genes and small molecules, and can be used to elucidate the mechanism of action of drugs. This area has recently attracted attention because of the prospect of a new generation of anti-cancer drugs.
The traditional method for identifying synthetically lethal interactions relied on mutant screens, typically in model organisms like Saccharomyces cerevisiae. With small genomes and rapid doubling times, these organisms were well-suited for the purpose. However, as researchers moved towards mammalian models, the efficiency of genomic perturbation became a major challenge. Horizon’s collection of cell lines with cancer-relevant mutations, combined with compound screens or loss-of-function screens with RNAi or CRISPR libraries, provide attractive model systems for performing synthetic lethal screens.
DLD-1 BRCA2 (-/-) cell line response to Olaparib
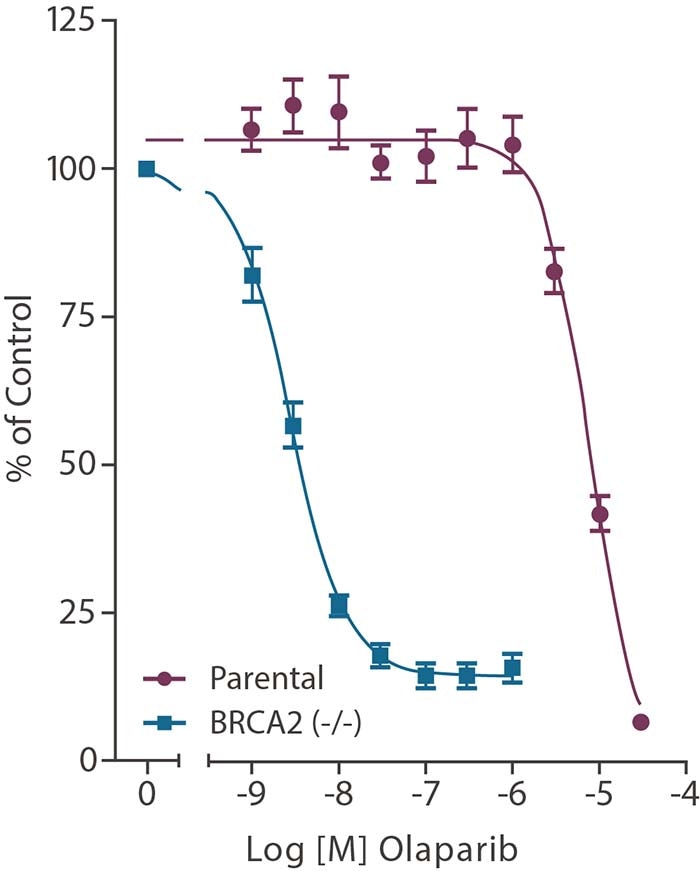
Figure 1. DLD-1 BRCA2 (-/-) cells show selective sensitivity to Olaparib over DLD-1 parental cells in a 10 day colony forming assay.
Synthetic lethality studies in BRCA2(-/-) cells
An example of synthetic lethality studies using knockout cell lines can be seen in figure 1, found in this application note. Briefly, a Human BRCA2(-/-) DLD-1 cell line was used as the basis for a small molecule screen and assessed for viability, with the goal of identifying compounds that inhibit a gene target that results in a lethal effect compared to DLD-1 BRCA2 (+/+) cells.
The tumor suppressor and DNA repair gene BRCA2 displayed synthetic lethality with another DNA repair enzyme, PARP. The use of isogenic pairs of cell line clones allowed the role of the BRCA2 mutation to be studied in isolation.
Compound screening in DNA repair cell lines
In figure 2, from a second application note, a selection of Horizon’s knockout cell lines were used to confirm involvement of particular DNA damage repair pathways in a compounds' mode of action and demonstrate opportunities for the exploitation of synthetic lethal interactions, using a 96 hour proliferation assay.
LIG4 and PRKDC (-/-) cell lines response to Etoposide
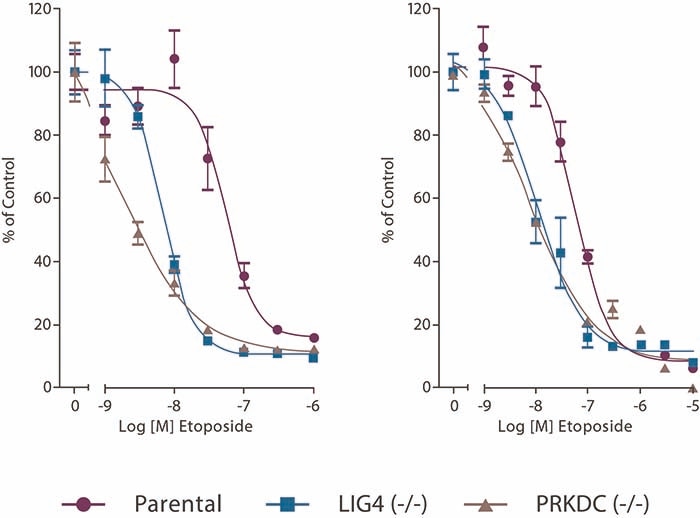
Figure 2. LIG4 and PRKDC (-/-) cells show selective sensitivity to Etoposide over HCT116 Parental cells in an 8 day colony forming assay and a 96 hour proliferation assay.
Order Products
Ready-made cell lines
Powerful cell line tools for applications such as pathway analysis or gene regulation assays to save researchers the time and resources of editing and characterizing clonal cell lines.
CRISPR guide RNA
Dharmacon Edit-R CRISPR guide RNAs are guaranteed to edit the target gene-of-interest, so you can be confident in your knockout result – see guarantee tab for details.
Cas9 nuclease
Purified and lentiviral expressed Cas9 nuclease solutions for CRISPR gene editing experiments
CRISPR controls
Ensure your system is optimized for successful CRISPR-Cas9 experiments
Helpful Resources
Read app note
Compound screening in BRCA2 (-/-) cell lines: a model for synthetic lethality
