カスタムPin-pointガイドRNAのご注文は、画面右側のツールをご利用ください。また、下記のガイドラインをご確認いただくか、PDF版をダウンロードしてご参照いただけます。詳しくは、Pin-point塩基編集プラットフォーム向けガイドRNA設計に関するアプリケーションノートをダウンロードしてご覧ください。
nCas9(NGG PAM配列)およびラット由来APOBECデアミナーゼに基づく、タンパク質ノックアウト実験用sgRNA選択ガイドライン
-
手動方法または公開されている塩基編集ガイドRNA設計ツール(参考:Example ResourcesセクションまたはAppendix A)を使用し、関心のある転写物に基づいて、目的のsgRNAプロトスペーサー(標的)配列の候補リストを作成してください。
- A) タンパク質ノックアウトを目的として、CAA、CAG、CGA、またはTGGコドンを終止コドンに変換可能なNGG PAMを有するsgRNAを設計し、早期終止コドンを生成します(図1)。この変換は、標的となるC塩基がスペーサー配列の5'→3'方向における3〜8番目の位置に存在する場合に可能です(図2)1。
- B) スプライス部位の破壊によるタンパク質ノックアウトの誘導
- NGG PAMを有するsgRNAの設計による、標的C塩基のスペーサー配列5'→3'方向における3〜8番目の位置への配置(図2参照)1
- 一般的なスプライスアクセプター部位:イントロン-AG|エクソン
- 一般的なスプライスドナー部位:エクソン|GT-イントロン
- GC塩基対からAT塩基対への変換によるスプライス部位の破壊2
-
必要に応じたsgRNA候補リストの絞り込み
- タンパク質ノックアウトの可能性が高いsgRNAの選定に向けた遺伝子構造の考慮
- 最初および最後のエクソンを標的とするsgRNAを回避する
- タンパク質の中央領域(トランスクリプトの中間10〜60%)を標的とするsgRNAを優先する。※追加候補が必要な場合は、60%以降のCDS領域も対象に含める
- タンパク質の必須ドメイン(例:受容体の膜貫通ドメイン)をコードするエクソンを標的とするsgRNAを優先する
- 非対称エクソンの標的化を優先する。※対称エクソンを標的とした場合、エクソンがスキップされても後続エクソンがフレーム内に保持される可能性あり3
- gRNAがすべての必要なアイソフォームを標的とすることを確認する
- 標的Cの5'側2塩基に基づくsgRNAの優先順位付け3
- 配列コンテキストがNTCまたはTCCの場合を優先する
- 配列コンテキストがNGCまたはGCCの場合を回避する

図1:シチジンのデアミネーションによって生成可能な終止コドン1

図2:塩基編集ウィンドウ。濃緑:高効率編集の可能性 淡緑:低効率編集の可能性 白:バイスタンダー編集の可能性
References
-
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5610906 (see figure 1)
-
https://www.nature.com/articles/s41467-021-22009-2 (see figure 1)
-
Poster - Performance and modularity of Horizon’s Pin-point™ base editing system characterized by arrayed and pooled screening platforms
Additional External Resources
-
BE-Designer http://www.rgenome.net/be-designer/
-
iSTOP https://www.ciccialab-database.com/istop/#/
-
PnB Designer https://fgcz-shiny.uzh.ch/PnBDesigner/
-
beditor https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6553823/ (requires the use of Python)
-
SpliceR https://pubmed.ncbi.nlm.nih.gov/33893286/
-
CRISPR-BETS https://zhangtaolab.org/software/crisprbets
すべてのカスタムsgRNA配列は、実験的な検証が必要です。これらのガイドラインおよび提案は、機能性を保証するものではありません。オフターゲット効果を最小限に抑えるため、sgRNA配列の特異性については常に考慮してください。
Revvityは本情報を「現状のまま」提供しており、受領者はこれをそのまま受け入れるものとします。受領者は、本情報の使用に関するすべての責任を負うものとし、第三者の知的財産権が関係する可能性についても自ら判断する責任を負います。
本書には、他のWebサイト、モバイルアプリケーション、またはインターネット上の場所(以下「第三者サイト」)へのリンクまたはアクセス手段が含まれる場合があります。これらのリンクは、あくまで便宜のために提供されるものであり、当社は第三者サイト、その内容、または第三者サイトを通じて提供される商品・サービスについて一切の管理・確認・責任を負いません。当社のプライバシー通知は第三者サイトには適用されず、第三者サイトに提供されたデータについては、利用者自身の責任で提供されるものとします。第三者サイトをご利用の際は、各サイトのプライバシーポリシーをご確認いただくことを推奨します。
Design and order custom Pin-point sgRNAs
New: Pin-point base editing sgRNA design tool
Use our new Pin-point base editing design tool to design and order custom sgRNAs for gene knockout.
Order Custom Pin-point sgRNAs
Pin-point base editing sgRNA ordering tool
Already know the 20nt DNA targeting sequence that you want to edit? Use our custom guide RNA ordering tool to enter an order for your custom Pin-point base editing sgRNAs.
Additional guidance for using the Pin-point base editing sgRNA design tool
Designing and ordering sgRNAs with the Pin-point base editing sgRNA ordering tool
Video walkthrough of how to use our new design tool
Pin-point base editing design tool user guide
Additional instructions for the use of the design tool
ガイドラインをPDFでダウンロード
Designing custom single guide RNAs (sgRNAs)
nCas9(NGG PAM配列)およびラット由来APOBECデアミナーゼに基づく、タンパク質ノックアウト実験用sgRNA選択ガイドライン
Need more help?
Contact Usラット由来のAPOBECデアミナーゼは、CをTに、またはGをAに変換するために使用できます。以下に両方の例を示します。
例1:Cを標的とする場合
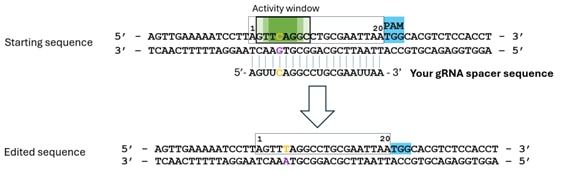
- 編集するヌクレオチドを特定します(オレンジ色で表示)。この例では、CAGがストップコドン(TAG)に変換されます。NGGのPAM配列は、常に標的鎖*のスペーサーの3'末端側に位置することを考慮してください。
- アクティビティウィンドウ内に配置されていることを確認します(濃い緑色ほど編集効率が高い可能性を示します)。
- スペーサー配列は非標的鎖に結合し、それに相補的である必要があります。
- 配列は5'から3'方向で、DNA配列として入力します(UではなくTを使用)。この例では、入力配列は「5'- AGTTCAGGCCTGCGAATTAA - 3'」となります。
例2:Gを標的とする場合
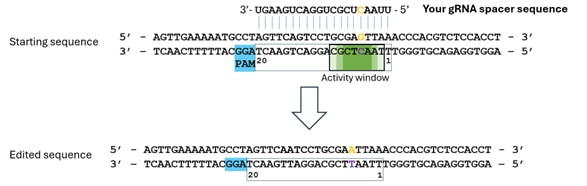
- 編集するヌクレオチドを特定します(オレンジ色で表示)。この例では、スプライスアクセプター部位(AG)が変異させられます。NGGのPAM配列は、常に標的鎖*のスペーサーの3'末端側に位置することを考慮してください。
- アクティビティウィンドウ内に配置されていることを確認します(濃い緑色ほど編集効率が高い可能性を示します)。
- スペーサー配列は非標的鎖に結合し、それに相補的である必要があります。
- 配列は5'から3'方向で、DNA配列として入力します(UではなくTを使用)。この例では、入力配列は「5'-TTAACTCGCTGGACTGAAGT-3'」となります。
*標的鎖 = デアミナーゼが作用するC基質が存在する鎖
