- Products
- カスタムオリゴ合成
- カスタムsiRNA合成
カスタムsiRNA合成
siRNAのカスタム合成がこれまでになく簡単になりました。
ご要件に見合うデザイン済みsiRNA製品が無い場合には、オンラインデザインツールと豊富な合成オプションを使用して、アプリケーションに固有のカスタムsiRNAをデザインしてください。修飾、量、および精製オプション等の様々な組み合わせをオンラインでデザイン/ご注文いただけます。 siRNAカスタムツールの使用方法に関するショートビデオをご覧ください。
カスタムsiRNA合成のご注文
修飾または非修飾siRNAの両方を注文いただけます。特許取得済みの化学修飾(ON-TARGET、ON-TARGETplus、siSTABLE、またはAccell)および追加のカスタム修飾が利用可能です。
カスタムSMARTpoolのデザインと化学合成
デザイン済みsiRNAコレクションを補完し、幅広いRNAi/実験をサポートするために、私共のsiRNA設計の専門チームは、ゲノムワイドのデザイン済み製品以外の遺伝子をターゲットとするカスタムSMARTpool試薬を提供できます。
siDESIGNセンター
標準ではない生物種の遺伝子、特定のスプライスバリアント、または遺伝子ファミリーや複数の生物種の相同な遺伝子領域をターゲットとするsiRNAの設計をご希望の場合は、siDESIGNセンターをご利用ください。
前臨床用またはOEM RNA/DNA合成
まとまった量のRNA/DNA合成の前臨床アプリケーション向けまたは受託製造(OEM)サービスをご希望の場合は、 こちらをご覧ください。
Alternatively, if you would prefer a custom generated quote fill out the custom synthesis quote form.
siRNA収量表
未修飾のsiRNAの場合、次のような概算収量が期待できます。
| Standard (A4) | HPLC purified | in vivo | in vivo HPLC | |||||
|---|---|---|---|---|---|---|---|---|
| nmol | mg | nmol | mg | nmol | mg | nmol | mg | |
| 0.025 µmol scale | 20 | 0.25 | - | - | - | - | - | - |
| 0.05 µmol scale | 40 | 0.5 | 20 | 0.25 | 25 | .3 | - | - |
| 0.2 µmol scale | 150 | 2 | 80 | 1 | 100 | 1.3 | 50 | 0.65 |
| 0.4 µmol scale | 300 | 4 | 160 | 2 | 200 | 2.6 | 100 | 1.3 |
| 1.0 µmol scale | 750 | 10 | 320 | 4 | 500 | 6.6 | 250 | 3.3 |
| 2.0 µmol scale | 1500 | 20 | 750 | 10 | 1000 | 13 | 500 | 6.6 |
| 5.0 µmol scale | 3750 | 50 | 1875 | 25 | 2500 | 33 | 1250 | 16 |
| 10.0 µmol scale | 7500 | 100 | 3750 | 50 | 5000 | 66 | 2500 | 33 |
専門家チームによるsiRNA設計支援が利用可能です。
当社には、RNAi研究を強化するためのさまざまな代替siRNA設計が備わっています。
ヒト、マウス、またはラットのモデルをご使用の場合は、デザイン済みsiRNA製品がすでに有る可能性があります。Websiteの上部の検索バーで遺伝子を検索してみてください。
当社の設計専門チームとSMARTpoolテクノロジーを活用してください。 安心してカスタマイズできるように、機能が保証されたカスタムSMARTpoolをお届けします。
私たちがサポートする代替siRNAデザインには以下が含まれます。
- ブラントエンド
- 非対称
- より長い二本鎖(23 nt以上)
- ミスマッチストランド
- 代替塩基またはリンケージ
必要なものが見つからない場合、オンライン注文では入手できない追加の仕様が必要な場合、見積もりをご請求ください
さらにサポートが必要な場合は、テクニカルサポートにお問い合わせください。
For more details on Revvity's continued compliance with the United States Framework For Nucleic Acid Synthesis Screening, please see our self-attestation.
様々なsiRNAソリューション(特異性、安定性、またはセルフデリバリー等)をご用意しています。
より性能が向上しているカスタムsiRNAをご使用いただくために、当社独自の化学修飾パターンを1つご選択ください。
独自仕様およびその他のすべてのsiRNA修飾は、カスタムsiRNA合成 リクエストを通じてご利用いただけます。
| ON-TARGET | ON-TARGETplus | Accell | siSTABLE | |
|---|---|---|---|---|
| RISCによるセンス(passenger)鎖の取り込みを阻害 | ✔ | ✔ | ✔ | ✔ |
| ターゲットへの特異性を高めるためのアンチセンス鎖seed領域の修飾 | ✔ | |||
| エンドヌクレアーゼおよびエキソヌクレアーゼによる分解に耐性 | ✔ | ✔ | ||
| トランスフェクション試薬不要で細胞へ導入 | ✔ | |||
| デザイン済みsiRNAとしても提供 | ✔ | ✔ |
ON-TARGET修飾により、アンチセンス鎖バイアスが保証されます。
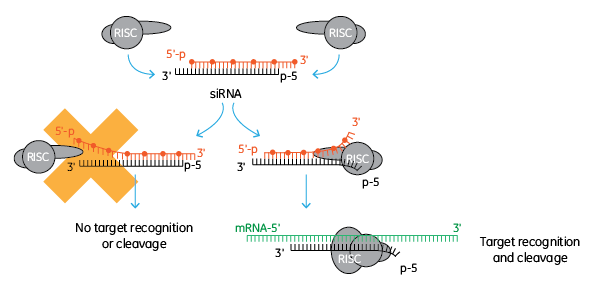
ON-TARGET修飾は、センス(passenger)鎖がRISCによって取り込まれるのをブロックすることにより、アンチセンス鎖のRISCへの取り込みを促進します。これは、アンチセンス(guide)鎖のRISCへの取り込みを確実にするための優れたな方法です。すべての独自のsiRNA修飾(ON-TARGETplus、Accell、siSTABLE)は、このセンス鎖修飾を含み、アンチセンス(ガイド)鎖によるノックダウンを促進します。
オフターゲットを減らすためのON-TARGETplus siRNA二本鎖修飾パターン
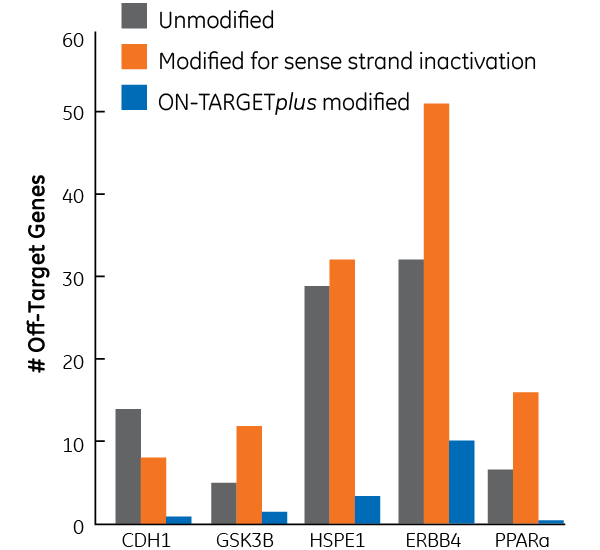
2006年の論文は、オフターゲット効果が主にアンチセンス鎖seed活性によって引き起こされることを示しています1。したがって、センス鎖の不活性化だけでは、オフターゲット遺伝子の総数は減少しません。ON-TARGETplus修飾は、siRNAの両方の鎖を考慮しています。
-
センス鎖は、RISCとの相互作用を妨げ、アンチセンス鎖のRISCへの取り込みを促進するように修飾されています。
-
アンチセンス鎖seed領域は、seed関連のオフターゲットを最小限に抑えるように修飾されています。
ON-TARGETplusの修飾パターンは、オフターゲットを劇的に減少します。図のsiRNAによって誘発されたオフターゲット効果は、マイクロアレイ分析を使用して定量化されました。各ターゲットに対して、3つの異なるsiRNAが使用されました(未修飾、センス鎖不活性化、およびON-TARGETplus修飾)。データは、2倍以上ダウンレギュレートされている遺伝子を表しています。HEK293細胞に0.2µLのDharmaFECT 1を使用して100 nM siRNAをトランスフェクトしました。データは24時間で分析しました。
1 Jackson, A.L. et al. "Position-specific Chemical Modification Increases Specificity of siRNA-mediated Gene Silencing." RNA 12.7 (2006) 1197-1205.
トランスフェクション試薬不要の心筋細胞におけるAccell siRNA導入と遺伝子ノックダウン
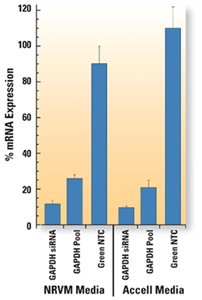
新生児ラット心室筋細胞を、1 µM Accell Green(A; Cat# D-001950-01)またはRed(B; Cat# D-001960-01)Non-targeting siRNAとともに、Accell delivery media(Cat# B-005000)で72時間インキュベートしました。核はDAPI(青)で染色されました。標識されたコントロールの取り込みは、ほぼすべての細胞でびまん性の細胞質局在を示しました。
棒グラフは、新生児ラット脳室筋細胞(NRVM)培地またはAccell delivery mediaを用いて、Accell GAPD Control siRNA(Cat#D-001930-03)およびPool(Cat#D-001930-30)コントロール試薬による遺伝ノックダウンを行った場合のGAPDH mRNA発現レベルを示しています。
筋細胞は、Maass AH&Buvoli M.心筋細胞の調製、培養、および遺伝子導入に記載されているように調製しました(Methods Mol Biol. 2007;366: 321-30.)。mRNA発現はQuantiGene branched DNAアッセイ(Panomics)によって決定しました。
siSTABLEで修飾されたsiRNAはヌクレアーゼによる分解に抵抗します。
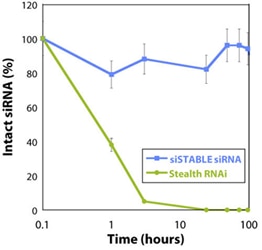
従来のsiRNAは血清を含む環境で数分以内に分解されるため、in vivoでのsiRNAの使用には問題がありました。このグラフは、siSTABLE修飾パターンが、Stealth RNAi(Invitrogen)と比較して、100%ヒト血清の存在下でsiRNAの半減期を劇的に延長することを示しています。
Accell siRNAには、これらの安定性を高める修飾が含まれているだけでなく、トランスフェクション試薬不要でトランスフェクションが困難な細胞にデリバリーすることができます。
| Fluorophore | λ max abs (nm) | λ max em (nm) | Comparable to | 5' or 3' | |
|---|---|---|---|---|---|
Fluorescein / 6-FAM
5'-Fluorescein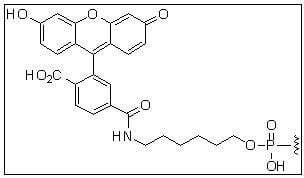
Description: Fluorescein is often used in fluorescence experiments to demonstrate the kinetics of folding or substrate binding. Fluorescein is also used as a donor to track optimal changes related to folding or substrate binding to intermolecular interactions.
Reference: Science 266: 785-789 (1994), EMBO J. 17: 2378-2391 (1998) |
494 | 520 | - | both* | |
TAMRA
5'-TAMRA-hexyl linker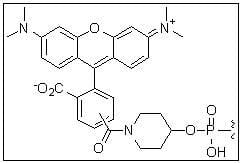
Description: TAMRA is a strongly absorbing dye with a wide variety of applications. This modification is coupled from the 5'- or 3'-end of an oligonucleotide to either the 5th or 6th position of the dye.
References: Nucl. Acids. Res. 24: 4535-4542 (1996), Biochem. 39: 14487-14484 (2000) |
565 | 580 | - | both* | |
Cy3
Modification Name: 5’-Cy3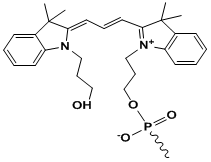
Modification Code: Cy3 Unit Molecular Weight: 507.59 g/mol Cy3 Extinction Coefficient: 136,000 Excitation/Emission Max: 547 nm/563 nm Unit Structure: Modification Name: 3’-Cy3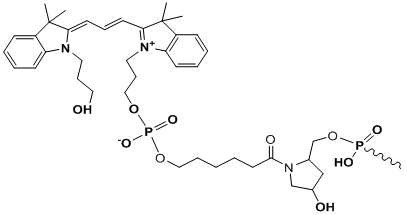
Modification Code: Cy3-3’ Unit Molecular Weight: 800.85 g/mol Cy3 Extinction Coefficient: 136,000 Excitation/Emission Max: 547 nm/563 nm Unit Structure: |
547 | 563 | - | both* | |
Cy5
Modification Name: 5’-Cy5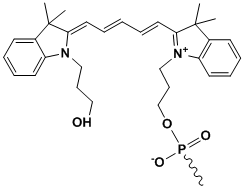
Modification Code: Cy5 Unit Molecular Weight: 533.63 g/mol Cy5 Extinction Coefficient: 250,000 Excitation/Emission Max: 646 nm/662 nm Unit Structure: Modification Name: 3’-Cy5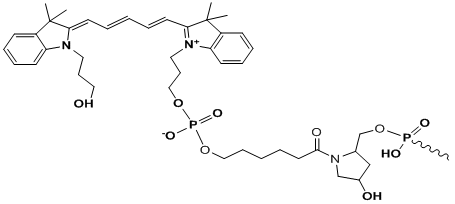
Modification Code: Cy5-3’ Unit Molecular Weight: 826.88 g/mol Cy5 Extinction Coefficient: 250,000 Excitation/Emission Max: 646 nm/662 nm Unit Structure: |
646 | 662 | - | both* | |
Cy5.5
Modification Name: 5’-Cy5.5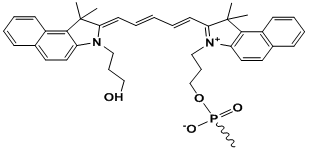
Modification Code: Cy5^5 Unit Molecular Weight: 633.75 g/mol Cy5.5 Extinction Coefficient: 209,000 Excitation/Emission Max: 688 nm/707 nm Unit Structure: Modification Name: 3’-Cy5.5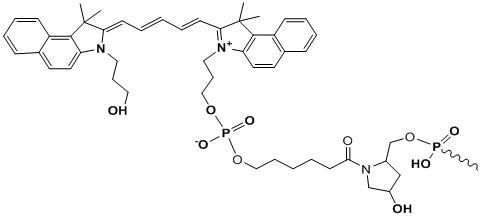
Modification Code: Cy5^5-3’ Unit Molecular Weight: 927.00 g/mol Cy5.5 Extinction Coefficient: 209,000 Excitation/Emission Max: 688 nm/707 nm Unit Structure: |
688 | 707 | - | both* | |
上記の表に記載されている色素オプションのいくつかは、他のオプションよりも高いオリゴ収量を得られます。アプリケーションに最適な色素ラベルの選択については、テクニカルサポートにお問い合わせください。実験に代替色素が必要になることがすでにわかっている場合は、オンラインで見積もりをリクエストすることもできます。
AlexaFluor®は、Invitrogen Corporationの登録商標です。
蛍光色素に加えて、siRNAと一本鎖RNAの両方に適用できる他の化学修飾の幅広いポートフォリオを提供しています。
オフターゲット、セルフデリバリー、ヌクレアーゼ耐性を低減するための特殊な修飾パターンについては、「独自開発のsiRNA」の項をご覧ください。
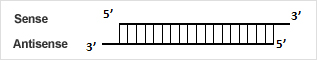
siRNAは、いずれの鎖のいずれの末端でも修飾できます。複数の修飾も可能ですが、可否はシーケンスまたは他の要因に依存する場合があります。詳細については">、テクニカルサポートにお問い合わせください。
特殊なカスタム合成アプリケーション向けに、さまざまなオリゴヌクレオチド修飾を提供します。オンライン注文の可否については、表をスクロールしてご確認ください。
NOTE: オンラインでご注文いただけない修飾もあります。サポートが必要な場合は、テクニカルサポートにお問い合わせください。
NOTE: 価格は米国における価格です。日本における価格はお見積りをご請求ください。
Scientific Support.
Filter by:
- siRNA
- ssRNA
- ssRNA-3'
- ssRNA-Internal
- ssRNA-5'
- miRNA
- SHOW ALL
Showing modifications available for:
- siRNA
- ssRNA
- ssRNA-3'
- ssRNA-Internal
- ssRNA-5'
- miRNA
- All
一本鎖RNAの標準的な修飾は、リクエストに応じてsiRNAに適用することもできます。siRNAへの追加の修飾やオプションについては 、 テクニカルサポート にお問い合わせいただ くか、 見積りをご請求ください。
| Standard RNA Bases | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
|---|---|---|---|---|---|---|
(A,C,G,U)
Standard RNA Bases (A, C, G, U)
Modification Code:
Unit Structure: 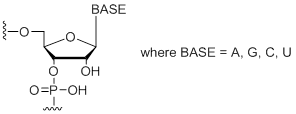
Unit Molecular Weight:
|
A,C,G,U | $6.63 | $8.36 | $13.97 | $18.36 | $28.15 |
| 2'-Omethyl RNA Bases | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
2'-OMe RNA Bases (A, C, G, U)
2'-OMe RNA Bases (A, C, G, U)
Modification Code:
Description:
Unit Structure: 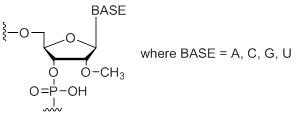
Unit Molecular Weight:
References:
|
mA,mC,mG,mU | $6.63 | $8.36 | $13.97 | $18.36 | $28.15 |
| Standard DNA Bases | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
2'-Deoxy Bases (A, C, G, T)
2'-Deoxy Bases (A, C, G, T)
Modification Code:
Description:
Unit Structure: 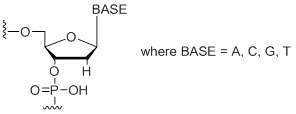
Unit Molecular Weight:
References:
|
dA,dC, dG, dT | $1.15 | $2.30 | $3.20 | $5.10 | $8.30 |
| Base Modifications | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
1-Methyl-guanosine
1-Methyl-guanosine
Modification Code:
Unit Structure: 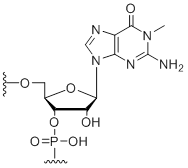
Unit Molecular Weight:
References:
|
m1G | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
2,6-Diaminopurine
2,6-Diaminopurine
Modification Code:
Description:
Unit Structure: 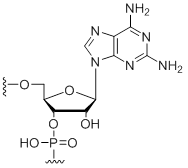
Unit Molecular Weight:
References:
|
DAP | $91.80 | $96.90 | $107.10 | $137.70 | $234.60 |
2-Methyl-adenosine
2-Methyl-adenosine
Modification Code:
Unit Structure: 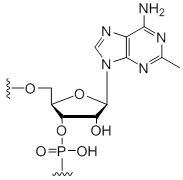
Unit Molecular Weight:
References:
|
m2A | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
2-Aminopurine
2-Aminopurine
Modification Code:
Description:
Unit Structure: 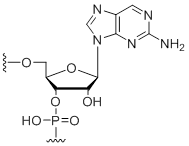
Unit Molecular Weight:
References:
|
2AP | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
4-Thio-uridine
4-Thio-uridine
Modification Code:
Description:
Unit Structure: 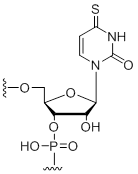
Unit Molecular Weight:
References:
|
4-S-U | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
5-Bromo-Uridine
5-Bromo-uridine
Modification Code:
Description:
Unit Structure: 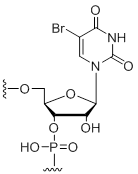
Unit Molecular Weight:
References:
|
U[5Br] | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Fluoro-cytidine
5-Fluoro-cytidine
Modification Code:
Description:
Unit Structure: 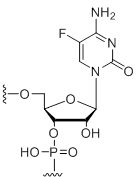
Unit Molecular Weight:
References:
|
C[5F] | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Fluoro-uridine
5-Fluoro-uridine
Modification Code:
Description:
Unit Structure: 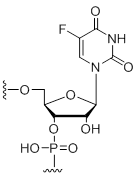
Unit Molecular Weight:
References:
|
U[5F] | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Iodo-uridine
5-Iodo-uridine
Modification Code:
Description:
Unit Structure: 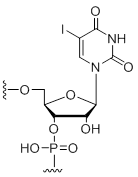
Unit Molecular Weight:
References:
|
U[5I] | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Methyl-cytidine
5-Methyl-cytidine
Modification Code:
Description:
Unit Structure: 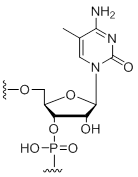
Unit Molecular Weight:
References:
|
5-M-C | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Methyl-Deoxycytidine
5-Methyl-deoxycytidine
Modification Code:
Description:
Unit Structure: 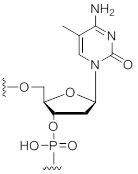
Unit Molecular Weight:
References: Freier, S. M. and Altmann, K.-H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically modified DNA:RNA duplexes. Nucleic Acid Res. 25, 4429-4443 (1997). |
5-M-dC | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Methyl-uridine
5-Methyl-uridine
Modification Code:
Description:
Unit Structure: 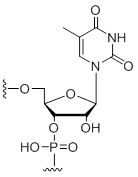
Unit Molecular Weight:
References:
|
rT | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
Inosine
Inosine
Modification Code:
Description:
Unit Structure: 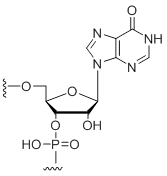
Unit Molecular Weight:
References:
|
I | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
N3-Methyl-uridine
N3-Methyl-uridine
Modification Code:
Unit Structure: 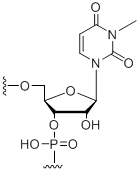
Unit Molecular Weight:
References:
|
3-M-U | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
N6,N6-Dimethyl-adenosine
N6,N6-Dimethyl-adenosine
Modification Code:
Description:
Unit Structure: 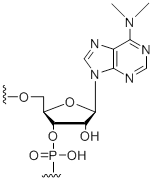
Unit Molecular Weight:
References:
|
DMA | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
N6-Methyl-adenosine
N6-Methyl-adenosine
Modification Code:
Description:
Unit Structure: 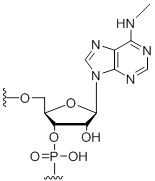
Unit Molecular Weight:
References:
|
m6A | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
Pseudouridine
Pseudouridine
Modification Code:
Description:
Unit Structure: 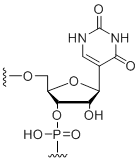
Unit Molecular Weight:
References:
|
~U | $91.80 | $96.90 | $107.10 | $137.70 | $234.60 |
Purine ribonucleoside
Purine ribonucleoside
Modification Code:
Description:
Unit Structure: 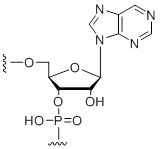
Unit Molecular Weight:
References:
|
Pu | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
Pyrrolo-cytidine
Pyrrolo-cytidine
Modification Code:
Description:
Unit Structure: 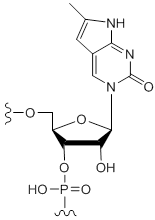
Unit Molecular Weight:
References:
|
pC | $91.80 | $96.90 | $107.10 | $137.70 | $234.60 |
Ribavirin
Ribavirin
Modification Code:
Description:
Unit Structure: 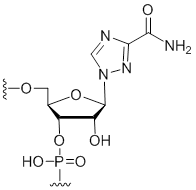
Unit Molecular Weight:
References:
|
RBV | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
| Backbone modifications | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
Phosphorothioate
Phosphorothioate
Modification Code:
Description:
Unit Structure: 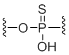
Unit Molecular Weight:
References:
|
* | $8.16 | $13.26 | $15.30 | $20.40 | $29.58 |
| Phosphorylation | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
5'-Phosphate
5'-Phosphate
Modification Code:
Unit Structure: 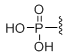
Unit Molecular Weight:
|
5'-P | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
| 2'-Modifications | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
2'-Amino-butyryl-pyrene-uridine
2'-Amino-butyryl-pyrene-uridine
Modification Code:
Description:
Unit Structure: 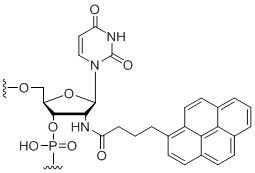
Unit Molecular Weight:
Pyrene Extinction Coefficient:
Excitation/Emission Max:
References:
|
2'-P-U | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
2'-Amino-cytidine
2'-Amino-cytidine
Modification Code:
Description:
Unit Structure: 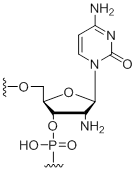
Unit Molecular Weight:
References:
|
2'-N-C | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
2'-Amino-uridine
2'-Amino-uridine
Modification Code:
Description:
Unit Structure: 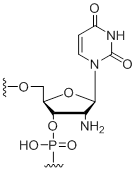
Unit Molecular Weight:
References:
|
2'-N-U | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
2'-Deoxy-uridine
2'-Deoxy-uridine
Modification Code:
Description:
Unit Structure: 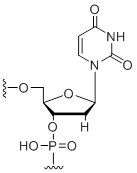
Unit Molecular Weight:
References:
|
dU | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
2'-Fluoro-adenosine
2'-Fluoro-adenosine
Modification Code:
Description: Unit Structure: 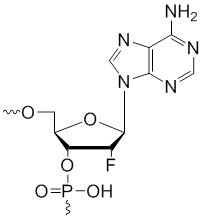
Unit Molecular Weight:
References: |
2'-F-A | $59.16 | $61.20 | $86.70 | $88.74 | $163.20 |
2'-Fluoro-cytidine
2'-Fluoro-cytidine
Modification Code:
Description:
Unit Structure: 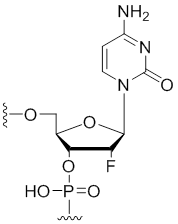
Unit Molecular Weight:
References:
|
2'-F-C | $40.80 | $43.86 | $58.14 | $66.30 | $102.00 |
2'-Fluoro-guanosine
2'-Fluoro-guanosine
Modification Code:
Description:
Unit Structure: 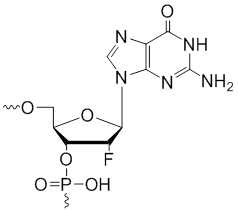
Unit Molecular Weight:
References:
|
2'-F-G | $59.16 | $61.20 | $86.70 | $88.74 | $163.20 |
2'-Fluoro-uridine
2'-Fluoro-uridine
Modification Code:
Description:
Unit Structure: 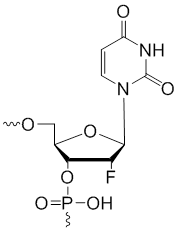
Unit Molecular Weight:
References:
|
2'-F-U | $40.80 | $43.86 | $58.14 | $66.30 | $102.00 |
2'-OMe-inosine
2'-OMe-inosine
Modification Code:
Description:
Unit Structure: 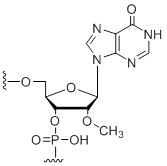
Unit Molecular Weight:
References:
|
mI | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
| Amino Modifiers | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
3'-Amino modifier C12
3'-Amino modifier C12
Modification Code:
Description:
Unit Structure: 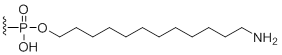
Unit Molecular Weight:
|
N12-3' | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
3'-Amino modifier C6
3'-Amino modifier C6
Modification Code:
Description:
Unit Structure: 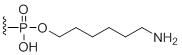
Unit Molecular Weight:
|
N6-3' | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
5'-Amino modifier C12
5'-Amino modifier C12
Modification Code:
Description:
Unit Structure: 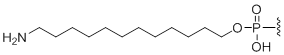
Unit Molecular Weight:
|
N12 | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
5'-Amino modifier C3
5'-Amino modifier C3
Modification Code:
Description:
Unit Structure: 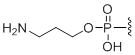
Unit Molecular Weight:
|
N3 | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5'-Amino modifier C5
5'-Amino modifier C5
Modification Code:
Description:
Unit Structure: 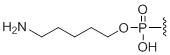
Unit Molecular Weight:
|
N5 | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5'-Amino modifier C6
5'-Amino modifier C6
Modification Code:
Description:
Unit Structure: 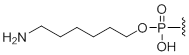
Unit Molecular Weight:
|
N6 | $74.46 | $81.60 | $91.80 | $117.30 | $204.00 |
5-Aminohexylacrylamino-uridine
5-Aminohexylacrylamino-uridine
Modification Code:
Description:
Unit Structure: 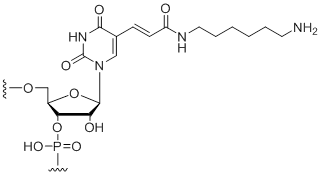
Unit Molecular Weight:
References:
|
5-LC-N-U | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
| Thiol Modifiers | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
3'-Disulfide thiol-modifier
3'-Disulfide thiol-modifier
Modification Code:
Description:
Unit Structure: 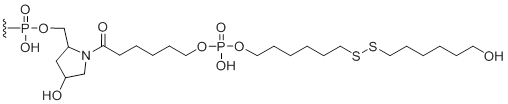
Unit Molecular Weight:
|
S-S-3' | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
5'-Disulfide thiol-modifier
5'-Disulfide thiol-modifier
Modification Code:
Description:
Unit Structure: 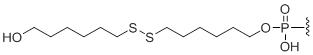
Unit Molecular Weight:
|
S-S | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
| Spacer Modifiers | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
C18 Spacer
C18 spacer
Modification Code:
Description:
Unit Structure: 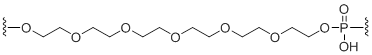
Unit Molecular Weight:
References:
|
18S | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
C3 Spacer
C3 spacer
Modification Code:
Description:
Unit Structure: 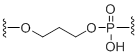
Unit Molecular Weight:
References:
|
C3 | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
C9 Spacer
C9 spacer
Modification Code:
Description:
Unit Structure: 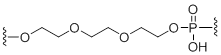
Unit Molecular Weight:
References:
|
9S | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
dSpacer
dSpacer
Modification Code:
Description:
Unit Structure: 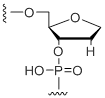
Unit Molecular Weight:
References: Vesnaver, G. et al. Influence of abasic and anucleosidic sites on the stability, conformation, and melting behavior of a DNA duplex: correlations of thermodynamic and structure data. Proc. Natl. Acad. Sci. USA 86, 3614-3618 (1989). Taniho, K. et al. Synthesis and biological properties of chemically modified siRNAs bearing 1-deoxy-D-ribofuranose in their 3'-overhang region. Bioorg. Med. Chem. Lett. 22, 2518-2521 (2012). |
dab | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
rSpacer
rSpacer
Modification Code:
Description:
Unit Structure: 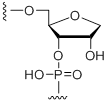
Unit Molecular Weight:
References: Vesnaver, G. et al. Influence of abasic and anucleosidic sites on the stability, conformation, and melting behavior of a DNA duplex: correlations of thermodynamic and structure data. Proc. Natl. Acad. Sci. USA 86, 3614-3618 (1989). Taniho, K. et al. Synthesis and biological properties of chemically modified siRNAs bearing 1-deoxy-D-ribofuranose in their 3'-overhang region. Bioorg. Med. Chem. Lett. 22, 2518-2521 (2012). |
rab | $229.50 | $284.58 | $321.30 | $397.80 | $703.80 |
| Chain Terminators | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
3' Inverted abasic
3' Inverted abasic
Modification Code:
Unit Structure: 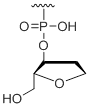
Unit Molecular Weight:
|
b | $45.90 | $53.04 | $64.26 | $79.56 | $135.70 |
3' Inverted deoxy-thymidine
3' Inverted deoxy-thymidine
Modification Code:
Description:
Unit Structure: 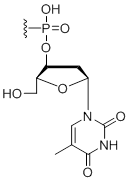
Unit Molecular Weight:
References:
|
idT-3' | $45.90 | $53.04 | $64.26 | $79.56 | $135.70 |
3'-Terminal 3'-Deoxy-Guanosine
3'-Terminal 3'-deoxy-guanosine
Modification Code:
Unit Structure: 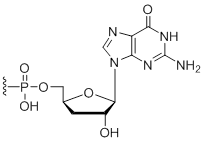
Unit Molecular Weight:
|
3'dG | $56.10 | $81.60 | $91.80 | $122.40 | $183.60 |
3'-Terminal dideoxy-cytidine
3'-Terminal dideoxy-cytidine
Modification Code:
Unit Structure: 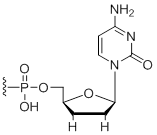
Unit Molecular Weight:
|
ddC | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
5' Inverted deoxy-thymidine
5' Inverted deoxy-thymidine
Modification Code:
Description:
Unit Structure: 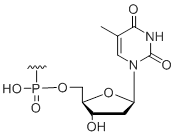
Unit Molecular Weight:
References:
|
idT | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
5' Terminal 5'-deoxy-ribo-adenosine
5' Terminal 5'-deoxy-ribo-adenosine
Modification Code:
Unit Structure: 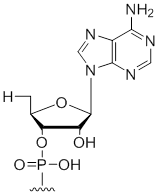
Unit Molecular Weight:
|
5'dA | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
5' Terminal 5'-deoxy-ribo-cytidine
5' Terminal 5'-deoxy-ribo-cytidine
Modification Code:
Unit Structure: 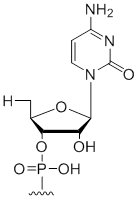
Unit Molecular Weight:
|
5'dC | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
5' Terminal 5'-deoxy-ribo-guanosine
5' Terminal 5'-deoxy-ribo-guanosine
Modification Code:
Unit Structure: 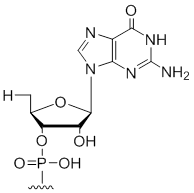
Unit Molecular Weight:
|
5'dG | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
5' Terminal 5'-deoxy-ribo-uridine
5' Terminal 5'-deoxy-ribo-uridine
Modification Code:
Unit Structure: 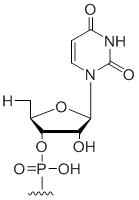
Unit Molecular Weight:
|
5'dU | $30.60 | $32.64 | $42.84 | $59.16 | $102.00 |
| Labeling | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
3-Biotin
3-Biotin
Modification Code:
Description:
Unit Structure: 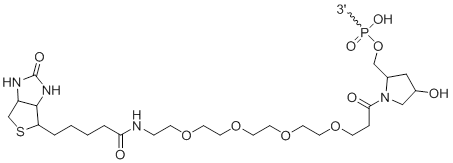
Unit Molecular Weight:
|
3'-Bi | $76.50 | $96.90 | $107.10 | $132.60 | $193.80 |
3'-Cholesterol
3'-Cholesterol
Modification Code:
Description:
Unit Structure: 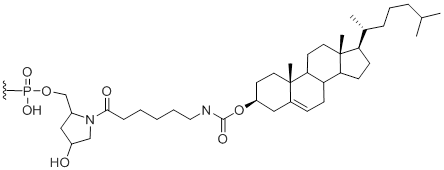
Unit Molecular Weight:
References:
|
3'-Chl | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
3' TEG-Cholesterol
3' TEG-Cholesterol
Modification Code:
Description:
Unit Structure: 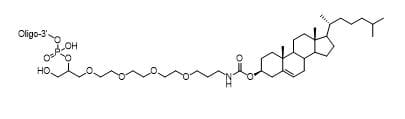
Chemical Formula:
Unit Molecular Weight:
References:
|
TEG-Chl-3' | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
3'-Cy3
3'-Cy3
Modification Code:
Description:
Unit Structure: 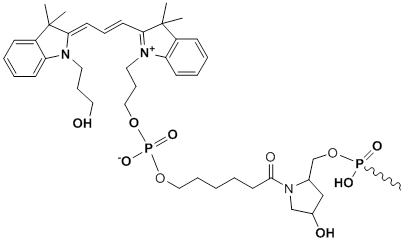
Unit Molecular Weight:
Cy3 Extinction Coefficient:
Excitation/Emission Max:
References:
|
Cy3-3' | $86.70 | $107.10 | $127.50 | $147.90 | $275.40 |
3'-Cy5
3'-Cy5
Modification Code:
Description:
Unit Structure: 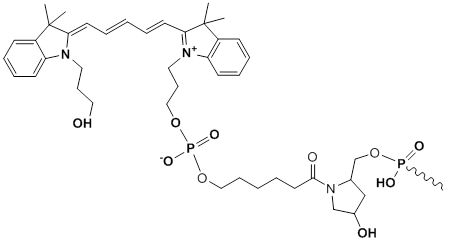
Unit Molecular Weight:
Cy5 Extinction Coefficient:
Excitation/Emission Max:
References:
|
Cy5-3' | $86.70 | $107.10 | $127.50 | $147.90 | $275.40 |
3'-Cy5.5
3'-Cy5.5
Modification Code:
Description:
Unit Structure: 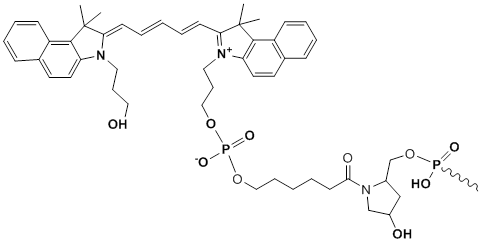
Unit Molecular Weight:
Cy5.5 Extinction Coefficient:
Excitation/Emission Max:
References:
|
Cy5^5-3' | $86.70 | $107.10 | $127.50 | $147.90 | $275.40 |
3’ Black Hole Quencher 1
3’ Black Hole Quencher 1
|
BHQ-1-3’ | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
3’ Black Hole Quencher 2
3’ Black Hole Quencher 2
|
BHQ-2-3’ | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
3'-Fluorescein
3'-Fluorescein
Modification Code:
Description:
Unit Structure: 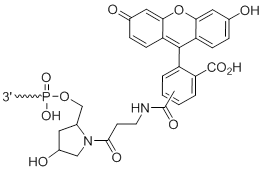
Unit Molecular Weight:
Extinction Coefficient:
Excitation/Emission Max:
|
3'-Fl | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
3'-Biotin LC
3'-Biotin LC
Modification Code:
Description:
Unit Structure: 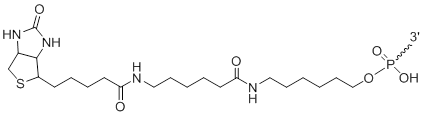
Unit Molecular Weight:
|
3'-LCBi | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
3'-Biotin LC LC
3'-Biotin LC LC
Modification Code:
Description:
Unit Structure: 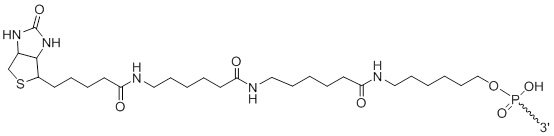
Unit Molecular Weight:
|
3'-2LCBi | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
3'-Puromycin
3'-Puromycin
Modification Code:
Description:
Unit Structure: 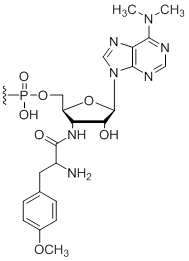
Unit Molecular Weight:
References:
|
Pmn | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
3'-TAMRA
3'-TAMRA
Modification Code:
Description:
Unit Structure: 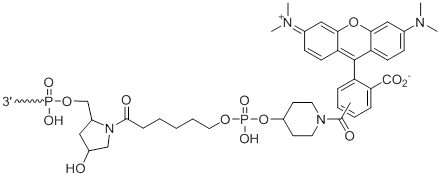
Unit Molecular Weight:
Extinction Coefficient:
Excitation/Emission Max:
|
3'-TAM | $242.76 | $285.60 | $336.60 | $387.60 | $673.20 |
5'-Biotin
5'-Biotin
Modification Code:
Description:
Unit Structure: 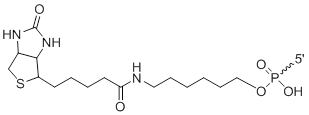
Unit Molecular Weight:
|
Bi | $91.80 | $96.90 | $107.10 | $158.10 | $275.40 |
5'-Cholesterol
5'-Cholesterol
Modification Code:
Description:
Unit Structure: 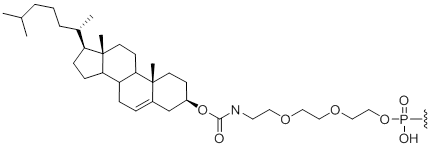
Unit Molecular Weight:
References:
|
Chl | $112.20 | $147.90 | $168.30 | $214.20 | $372.30 |
5' Cholesterol-TEG
5' Cholesterol-TEG
Modification Code:
Description:
Unit Structure: 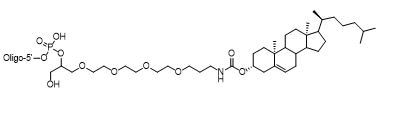
Chemical Formula:
Unit Molecular Weight:
References:
|
Chl-TEG | $336.60 | $443.70 | $504.90 | $664.60 | $1116.90 |
5'-Cy3
5'-Cy3
Modification Code:
Description:
Unit Structure: 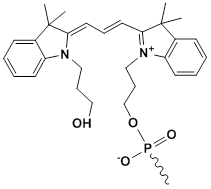
Unit Molecular Weight:
Cy3 Extinction Coefficient:
Excitation/Emission Max:
|
Cy3 | $76.50 | $96.90 | $117.30 | $137.70 | $255.00 |
5'-Cy5
5'-Cy5
Modification Code:
Description:
Unit Structure: 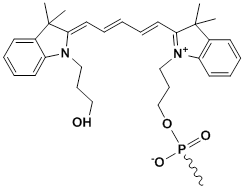
Unit Molecular Weight:
Cy5 Extinction Coefficient:
Excitation/Emission Max:
References:
|
Cy5 | $76.50 | $96.90 | $117.30 | $137.70 | $255.00 |
5'-Cy5.5
5'-Cy5.5
Modification Code:
Description:
Unit Structure: 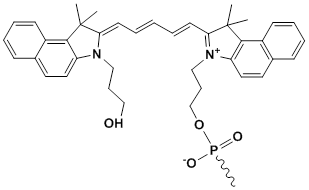
Unit Molecular Weight:
Cy5.5 Extinction Coefficient:
Excitation/Emission Max:
References:
|
Cy5^5 | $150.96 | $178.50 | $209.10 | $255.00/td> | $459.00 |
5’ Black Hole Quencher 1
5’ Black Hole Quencher 1
|
BHQ-1 | - | $650.76 | $740.52 | $943.50 | $1641.48 |
5’ Black Hole Quencher 2
5’ Black Hole Quencher 2
|
BHQ-2 | - | $650.76 | $740.52 | $943.50 | $1641.48 |
5'-Dabcyl
5'-Dabcyl
Modification Code:
Description:
Unit Structure: 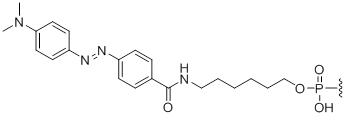
Unit Molecular Weight:
References:
|
Dabcyl | $102.00 | $132.60 | $147.90 | $183.60 | $326.40 |
5'-Fluorescein
5'-Fluorescein
Modification Code:
Description:
Unit Structure: 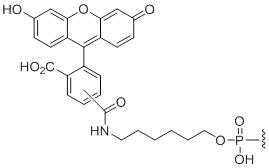
Unit Molecular Weight:
Extinction Coefficient:
Excitation/Emission Max:
|
Fl | $91.80 | $96.90 | $107.10 | $137.70 | $234.60 |
5'-Pyrene
5'-Pyrene
Modification Code:
Description:
Unit Structure: 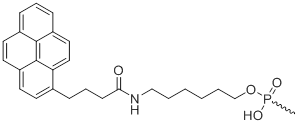
Unit Molecular Weight:
Pyrene Extinction Coefficient:
Excitation/Emission Max:
References:
|
5'-Pyr | $117.30 | $158.10 | $193.80 | $244.80 | $469.20 |
5'-TAMRA
5'-TAMRA
Modification Code:
Description:
Unit Structure: 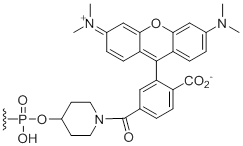
Unit Molecular Weight:
Extinction Coefficient:
Excitation/Emission Max:
|
TAM | $242.76 | $285.60 | $336.60 | $387.60 | $673.20 |
5'-TET
5'-TET
Modification Code:
Description:
Unit Structure: 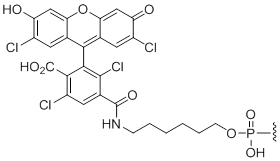
Unit Molecular Weight:
Extinction Coefficient:
Excitation/Emission Max:
|
TET | $76.50 | $94.86 | $107.10 | $132.60 | $234.60 |
| Degenerate Bases | Short Code | 0.05 µmol | 0.2 µmol | 0.4 µmol | 1.0 µmol | 2.0 µmol |
| Pricing shown is for first degenerate base coupling, subsequent additions at standard base pricing | ||||||
dN
dN
Modification Code:
Description:
Unit Structure: 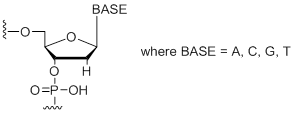
Unit Molecular Weight:
|
dN | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
dS
dS
Modification Code:
Description:
Unit Structure: 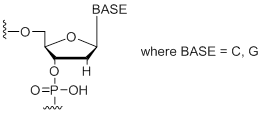
Unit Molecular Weight:
|
dS | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
dW
dW
Modification Code:
Description:
Unit Structure: 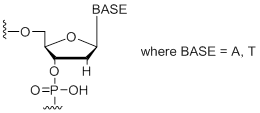
Unit Molecular Weight:
|
dW | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
mN
mN
Modification Code:
Description:
Unit Structure: 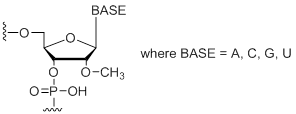
Unit Molecular Weight:
|
mN | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
rN
rN
Modification Code:
Description:
Unit Structure: 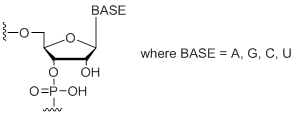
Unit Molecular Weight:
|
rN | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
rS
rS
Modification Code:
Description:
Unit Structure: 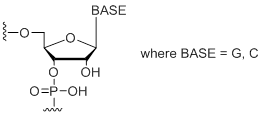
Unit Molecular Weight:
|
rS | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
rW
rW
Modification Code:
Description:
Unit Structure: 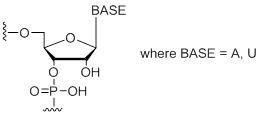
Unit Molecular Weight:
|
rW | $45.90 | $53.04 | $64.26 | $79.56 | $137.70 |
精製を追加しなくても、未修飾のsiRNA鎖の純度は通常80〜85%に達します。
次の場合は、化学合成siRNAの精製が推奨される場合があります。
- 3’末端または内部で化学的に修飾されている場合
- 同じ鎖で複数の修飾が施されている場合
- 高感度アッセイまたはin vivoアプリケーションでの使用を目的とする場合
未精製材料の精製または純度推定に関する推奨事項については、テクニカルサポートにお問い合わせください。
siRNA処理オプション:
| Single Strands (A1) | Standard (A4) | HPLC | In vivo | In vivo HPLC | |
|---|---|---|---|---|---|
| Desalted | ✔ | ✔ | ✔ | ✔ | |
| Deprotected | ✔ | ✔ | ✔ | ✔ | |
| Duplexed | ✔ | ✔ | ✔ | ✔ | |
| Purified | ✔ | ✔ | |||
| Endotoxin tested | ✔ | ✔ | |||
| Sodium counter-ion exchange | ✔ | ✔ | |||
| Recommended for modified siRNA (dyes, etc.) | ✔ | ✔ | |||
| Recommended for in vivo use | ✔ | ✔ | |||
| 2'ACE protected single-strands | ✔ |
siRNA処理について
- Desalted(脱塩):siRNA二本鎖はエタノール沈殿またはC18カラム脱塩のいずれかによって脱塩されています。
- Deprotected(脱保護):RNA塩基の2′-ACE保護基が除去(脱保護)されています。
- Duplexed(二本鎖):2つの相補的なsiRNA鎖がアニーリングされ、二本鎖が形成されています。
- Single-strand(一本鎖)(A1):siRNAは、センス鎖とアンチセンス鎖が別々のチューブで提供されます。個々のストランドは脱塩または脱保護されていません。
- Standard(標準)(A4):siRNAは脱塩および脱保護された二本鎖として提供され、再懸濁後にすぐに使用できます。
- HPLC:siRNA二本鎖は精製のためにイオン交換高速液体クロマトグラフィーを受けています。
- in vivo:siRNA二本鎖は、対イオン(Na +)交換、脱塩、滅菌ろ過、およびエンドトキシンテストによって処理されています。
- in vivo HPLC:二本鎖はin vivoプロセッシングとHPLC精製の両方を受けています。
RNAiテクノロジーの業界リーダーと提携して、最高品質のin vivo対応RNAi試薬をご利用ください。
Horizonは、皆様のin vivo実験が確実に成功することを願っています。実験計画を支援するために、次のオプションとガイダンスを提供しています。
in vitroでのsiRNAの評価を推奨します。
- 動物モデルを使用した費用のかかる実験を開始する前に、in vitro実験で高機能のsiRNAデザインを特定することをお勧めします。
- デザイン済みの4つのsiRNA試薬のセットは、複数のsiRNA配列をテストするのに理想的です。
- Individual:ヒト、マウス、ラットのすべての遺伝子に対して配列デザインの異なる4種類のsiRNAをご用意しています。4種類からいずれかのsiRNAをご注文ください。
- SMARTpool:1つの遺伝子に対して配列デザインの異なる4種類のsiRNAを1つのチューブにプールし、ご提供する製品です。
- Set of 4:4個のsiRNAを、個別のチューブに入れ、チューブ4本でセットとした製品です。
ヌクレアーゼ耐性を強化するための独自のsiRNA修飾パターンを検討してください。
- siSTABLE修飾は、エキソヌクレアーゼおよびエンドヌクレアーゼによる分解を防ぎます。siSTABLEは、siRNAが動物血清などのヌクレアーゼが豊富な生物学的環境にさらされる場合に推奨されます。
- Accell修飾は、安定性の強化に加えて、導入を強化する独自技術が組み込まれています。ご研究対象の細胞または組織タイプが標準的な導入モードに適していない場合は、Accellをお勧めします。
- in vivo 実験にsiSTABLEおよびAccell修飾siRNAを使用した最新の論文リストを参照してください。
- siSTABLEまたはAccellをカスタムsiRNA注文する。
必要なsiRNAの総量を注意深く計算します。
- 各コホートの動物の数、必要な処理法または用量の数、および各用量に必要なsiRNAの量を考慮してください。
- in vivo実験は通常、大量のsiRNAを必要とします。合成規模は、オンラインでは最大100mgに対応できます。それ以上の場合には、お問い合わせよりご要望をお知らせください(最大10gまで対応可能)。
- nmolからmgへの変換は、siRNA収量表をご参照ください。
動物への毒性が懸念される場合は、siRNAのin vivoプロセシングを検討してください。
- 対イオン(Na +)交換、滅菌ろ過、脱塩、エンドトキシンテストなどの特殊な合成後siRNA処理手順
- HPLC精製の有無にかかわらず利用できます。詳細については、siRNA処理オプションをご確認ください。
- in vivo実験の重要な考慮事項の詳細については、テクニカルノート「 in vivo RNAi: Biodistribution, Delivery, and Applications"」を参照してください。
