Arrayed crRNA libraries allow for functional genetic screens of genes with roles in different biological processes
Predesigned crRNA libraries enable rapid analysis of editing events in hundreds of genes or sites at once, but the identification of relevant hits and successful screening outcomes from these types of assays depends on transfection that has been optimized to have high efficiency and low toxicity. Transfection conditions that induce changes in cell viability and/or provide insufficient delivery of the targeting agent can mislead researchers toward false interpretations of data, which in the long run, are time-consuming and costly.
crRNA:tracrRNA delivery can be optimized with reverse transfection method
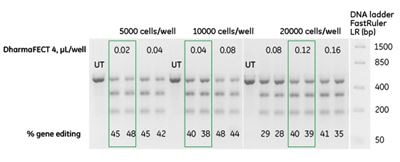
Forward transfection is a widely used method that works well for adherent cell types; however, if one is using suspension cells or a high-throughput format, reverse transfection, where cells are added to the plated transfection reagent complex, is a more appropriate approach. This method can be used to increase the throughput and reproducibility of a CRISPR-Cas9 screen by optimizing gene editing efficiency as demonstrated in our recently published application note: “Optimization of reverse transfection of Dharmacon Edit-R synthetic crRNA and tracrRNA components with DharmaFECT transfection reagent in a Cas9-expressing cell line.”
An un-cleavable ubiquitin moiety fused to EGFP allows constitutive degradation of the EGFP protein, while disruption of the proteasome components by functional protein knockout leads to accumulation of EGFP and detectable fluorescence. This functional knockout data shows that determining optimal transfection conditions prior to arrayed screening leads to high gene editing efficiency with minimal effect on cell viability.
Optimization of reverse transfection for arrayed crRNA
The following is a general workflow for reverse transfection of arrayed crRNA libraries using stable Cas9-expressing mammalian cells. Find more details in the full protocol for “Reverse Transfection of Arrayed crRNA”.
- Transfer working crRNA solution to each well of a 96-well V-bottom transfection mixture plate.
- Resuspend Edit-R tracrRNA to 10 μM stock solution in nuclease-free, 10 mM Tris pH 7.4 buffer.
- Prepare tracrRNA working solution by adding tracrRNA stock solution to serum-free medium.
- Add working tracrRNA solution to each well of 96-well V-bottom transfection mixture plate containing crRNA, prepared in step 1.
- Prepare transfection reagent working solution by diluting the transfection reagent stock solution in serum-free medium.
- Add transfection reagent working solution to each well of 96-well V-bottom transfection mixture plate containing the crRNA:tracrRNA complex.
- Immediately mix by pipetting gently up and down and incubate for 20 minutes at room temperature.
- Trypsinize cells, spin down, remove medium and resuspend in cell growth medium to a density determined optimal for your transfection.
- Add the transfection mixture to each well of three 96-well tissue culture plates.
- Add the cell suspension prepared in step 8 to each well of the three 96-well tissue culture plates.
- Incubate transfected cells at 37 °C in a humidified CO2 incubator for 48-72 hours before proceeding with the phenotypic assay or gene editing analysis.
Note: Optimal plating density will depend on growth characteristics of specific cell lines and assay requirements and these parameters should be determined experimentally. Exact parameters for crRNA:tracrRNA transfection in your cells of interest should be empirically determined through careful optimization prior to experimentation. Catalog crRNA library plates are supplied with columns 1 and 12 empty to allow addition of researcher-defined controls. We suggest including the following controls:
- Untreated cells
- Positive control crRNA
- Negative control: non-targeting crRNA
Order Products
DharmaFECT Transfection Reagents
For reliable transfection of small RNA and/or plasmids into any cell type
Helpful Resources
Optimization of reverse transfection of Edit-R synthetic crRNA and tracrRNA components - Application Notes
Maximize success of your arrayed synthetic crRNA knockout screen with careful transfection optimization, editing efficiency and phenotypic readout
Edit-R crRNA Libraries Reverse Transfection - Protocol
This is a reverse transfection protocol for crRNAs in arrayed plates
