- Non-mammalian research tools
- Yeast Parental Strains
Yeast Parental Strains

The Saccharomyces Genome Deletion Project (see Reference 1) has developed a unique collection of knock-out strains covering 96% of the yeast genome. This collection of over 6,000 gene-disruption mutants† provides a unique tool for the functional analysis of the yeast genome. The inclusion of distinct tags - or "molecular barcodes" - that identify each strain allows phenotypic analysis to be performed on a single gene basis or a genome-wide scale (see Reference 2). By using the yeast knock out strains for functional profiling, one can also gather much information about human gene function by analogy. Open Biosystems brings you access to the Yeast Knock-Out strains (YKO), a valuable resource for understanding gene function.
Highlights
- 6,000 individual genes are represented in four backgrounds, creating over 20,000 distinct knock out strains
- Molecular barcodes allow for parallel analysis of gene function
- Analyze individually by sequencing, or using whole genome approaches
- Whole gene deletions ensure loss-of-function
| Strain | Background | Genotype |
|---|---|---|
| MATa | BY4730 | MATa leu2Δ0 met15Δ0 ura3Δ0 |
| MATalpha | BY4739 | MATalpha leu2Δ0 lys2Δ0 ura3Δ0 |
| MATa | BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| MATalpha | BY4742 | MATalpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| Het/Hom Diploid | BY4743 | 4741/4742 |
| Homozygous diploids are in the BY4743 background unless 4730/4739 is indicated | ||
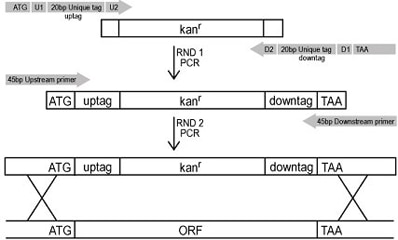
By means of two sequential PCR reactions - the first to incorporate the appropriate tags and confer the antibiotic resistance gene and the second to incorporate the mitotic recombination sites - each ORF was replaced with a KanMX cassette using homologous recombination. This method allowed for greater than 95% of the ORFs to be knocked-out.
Start screening sooner with yeast knockouts
Evidence for the functional role of a gene can be obtained by analysis of the phenotypic changes exhibited by each mutant strain under a given condition. While this type of analysis has been performed in the past using traditional genetic screens or random mutagenesis, the yeast knock out strains offer advantages over both of these methods.
One advantage is that the phenotypes of the yeast mutant strains reflect a complete loss of function of the given gene, where this is not assured using classical methods. Furthermore, in contrast to traditional screening, the gene identification is known a priori, thereby removing the time-consuming task of determining the responsible gene.
Also eliminated is the time and tediousness involved in creating a single yeast knock out strain. A single knock out strain is already available for nearly all of the yeast genes in the genome, so all you have to do is request the strain of interest.
Molecular Barcodes Facilitate Whole Genome Approaches
The mutant yeast strains can also be analyzed individually via sequencing or by using a whole genome approach. The molecular barcodes allow for parallel analysis of gene function using oligonucleotide arrays. The fitness of a given strain under a specific condition can be determined by calculating the abundance of that strain's DNA. This is achieved using an oligonucleotide array printed with the complementary tag sequence necessary to detect the presence of a molecular barcode (see References 1 and 2).
A wealth of information - including genetic and physical maps along with DNA sequence information - is available from the Saccharomyces Genome Database (SGD) at Stanford University.
Footnotes
† While >6,000 represents the number of individual genes, there are four backgrounds (haploids of MATa and MATalpha, heterozygous diploid, and homozygous diploid) with many genes available in more than one background, thereby creating over 20,000 knock out strains.
Disclaimer
We provide certain clone resources developed by leading academic laboratories. Many of these resources address the needs of specialized research communities not served by other commercial entities. In order to provide these as a public resource, we depend on the contributing academic laboratories for quality control. Therefore, these are distributed in the format provided by the contributing institution "as is" with no additional product validation or guarantee. We are not responsible for any errors or performance issues. Additional information can be found in the product manual as well as in associated published articles (if available). Alternatively, the source academic institution can be contacted directly for troubleshooting.
Shipping Information
Individual yeast knock out strains are provided as a live culture in a 2 mL tube. Each tube contains YPD broth with G418 (200 µg/mL) and is supplemented with 15% glycerol. Within 3 to 4 days of receiving your order we will ship your clone at room temperature via express delivery. Store the stock clone at 4°C for up to one week or -80°C indefinitely.
The Yeast Knock Out collections are available as frozen glycerol stocks in 96-well plates. The glycerol stocks are a culture of the Yeast Knock Out in YPD media and glycerol. These plates ship on dry ice and can be maintained indefinitely at -80°C.
Contact Scientific Support for a quote and estimated shipping time on bulk orders.

- E. A. Winzeler et al., Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science. 285(5429), 901-906 (6 August 1999).
- G. Giaever et al., Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418, 387-391 (2002).
- A. Wach, A. Brachat, R. Poehlmann, P. Philippsen, New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10(13), 1793-1808 (December 1994).
Product data
Product inserts
-
Trans-Lentiviral shRNA Packaging Kit - Product Insert
-
Yeast Parental Strain BY4730 - Product Insert
-
Yeast Parental Strain BY4739 - Product Insert
-
Yeast Parental Strain BY4741 - Product Insert
-
Yeast Parental Strain BY4742 - Product Insert
-
Yeast Parental Strain BY4743 - Product Insert
-
Yeast S288C Strain - Product Insert
-
Yeast W303 Heterozygous Diploid Strain - Product Insert
-
Yeast W303 MATa Strain - Product Insert
-
Yeast W303 MATalpha Strain - Product Insert
