- Dharmaconスクリーニングライブラリー
- Edit-R lentiviral sgRNAプール化ライブラリー
Edit-R lentiviral sgRNAプール化スクリーニング用ライブラリー
アルゴリズムで最適化されたガイドRNAでライブラリーの特異性を確保します。細胞あたり1種類のsgRNA配列の発現で効率的なゲノム編集が行えます。
ノックアウトスクリーニング用の全ゲノム、druggableゲノム、遺伝子ファミリー、パスウェイライブラリー
注意事項:プール化ライブラリーは、新しく改良されたベクター・スキャフォールドで更新されました。ご不明な点がございましたら、Scientific supportまでお問い合わせください。


複数の自動化機器を導入せずに、偏りのない表現型のノックアウトスクリーニングを実行します。
プール化レンチウイルスライブラリーを用いたスクリーニングでは、DNAレベルで数百、さらには数千の遺伝子をノックアウトします。
CRISPRガイドRNAが標的DNAの二本鎖切断を引き起こし、タンパク質の機能的破壊を引き起こす能力は、ガイドRNA(gRNA)の配列と標的遺伝子内の標的位置によって異なります。これに対処するために、Dharmacon 試薬は、アルゴリズムを使用して、ヒトゲノムおよびマウスゲノムに対して高度に特異的で機能的なRNAガイドの設計をしています。
Edit-R CRISPR RNAアルゴリズムは、次のようなガイドRNA配列を生成します。
- 複数のミスマッチやギャップのあるアラインメントの検出を含む、迅速かつ徹底的な特異性分析を備えた最適化されたアラインメントプログラム
- 単なるindelの生成ではなく、遺伝子の機能的破壊に関連する特性を特定するための体系的な評価研究(1100を超えるガイドRNAを使用)により、標的遺伝子の機能的ノックアウトが増加
- New! Edit-RヒトsgRNAデザインは2025年最新版RefSeqに更新されました。これにより、効率的なタンパク質ノックアウトを実現するための、最も特異的でゲノム関連性の高いガイドが提供されます。この更新により、Edit-Rアルゴリズムは最新のゲノムアノテーションをより正確かつ効率的に標的とすることが可能となり、ご研究ニーズに最適なソリューションを提供します。ご質問がございましたら、Scientific Supportまでお問い合わせください。
Highlights:
- Cas9発現用のレンチウイルスベクターとsgRNA発現用の遺伝子特異的ベクターを利用した効率的な2ベクターシステム
- 合理的に設計されたEdit-RレンチウイルスsgRNAによる、機能的に検証された独自のアルゴリズムを使用して比類のない特異性を備えた効率的な遺伝子ノックアウト
- ヒットの信頼性を高めるために、whole genomeライブラリーの場合、ヒトゲノム全体で遺伝子あたり 4あるいは8個の sgRNA、マウスゲノム全体では遺伝子あたり5~10個のsgRNAで深く広範囲にカバー
- Edit-R Lentiviral Cas9 Nuclease Reagentsのプロモーターの柔軟性と組み合わせて、生物学的に関連する細胞タイプで強力な編集を行うことができます。
新しいEdit-R Human Lentiviral sgRNAプール化スクリーニング用ライブラリーには、プールごとに以下のものが含まれます。
- ≥ 5 x 108 TU/mL ( ± 20%) レンチウイルス粒子の10 x 100 µL パックサイズ
- NCBI Reference Sequence Databaseのタンパク質をコードする遺伝子を標的とする、遺伝子あたり4あるいは8個のsgRNA(全ゲノム)および最大8個のsgRNA(遺伝子ファミリーおよびパスウェイライブラリー)
- 最大100個のnon-targeting sgRNA陰性コントロールは、ヒトゲノムのどの遺伝子ともアライメントしない(標的としない)ことがバイオインフォマティクスにより確認されています。
- 遺伝子ファミリーおよびパスウェイライブラリーには、最大34のタンパク質コード遺伝子を標的とする最大340の遺伝子特異的sgRNA陽性コントロールが含まれます(各遺伝子につき10 sgRNA)。一般的な参照遺伝子 (ACTB、GAPDH、LAMB1、PPIB)を含みます。
- 遺伝子アノテーション、sgRNA標的配列、コントロールの完全なリスト、およびマッッピングされた数百万リードあたりのカウントを含む、完全なライブラリー情報を含むデータファイル
Edit-R Mouse Lentiviral sgRNAプール化スクリーニング用ライブラリーには、プールごとに以下のものが含まれます。
- ≥ 5 x 108 TU/mL ( ± 20%) のレンチウイルス粒子の分注済みチューブ
- 5000以下のライブラリーコンストラクトの場合、8 x 25 µL(合計200 µL)
- 5000以上のライブラリーコンストラクトの場合、16 x 25 µL(合計400 µL)
- NCBI Reference Sequence Databaseのタンパク質をコードする遺伝子を標的とする遺伝子あたり5~10個のsgRNA
- 100のnon-targeting sgRNAネガティブコントロールは、ヒト、マウスのゲノムのどの遺伝子とも一致しない(標的にしない)ことがバイオインフォマティクスで確認されています。
- 最大34のタンパク質コード遺伝子(遺伝子あたり10 sgRNA)を標的とする最大340の遺伝子特異的sgRNAポジティブコントロール。一般的なリファレンス遺伝子(ACTB、GAPDH、LAMB1、およびPPIB)を含みます。
- 完全なライブラリー情報を含むデータファイル(含:遺伝子注釈、sgRNA標的配列、コントロールの完全なリスト、およびマップされたリードの数百万あたりのカウント)
当社の検証済みプロトコルで推奨される製品:
- ゲノム編集の検証および実験の最適化のための、ベクターに適合したEdit-Rレンチウイルス陽性およびnon-targeting sgRNAコントロール
- 目的の細胞における相対的導入効率の決定および導入条件の最適化のための、最適プロモーターおよび蛍光レポーター(TurboGFPまたはTurboRFP)を有するSMARTvector Non-targetingコントロールパーティクル
- Edit-R Lentiviral Cas9 ヌクレアーゼ試薬またはCas9安定細胞株
- NEW V2バックボーンのヒトライブラリー:
- 新製品: NGSライブラリー調製キット(推奨)
- プライマーセットオプションは12または24の全ゲノムスクリーニングに対応した2種類のサイズから選択可能
- gDNAからsgRNAコンストラクトを偏りなく増幅およびシーケンスするようにデザイン
- 加試薬を必要とせず、Illumina®プラットフォームでのシーケンシングに対応する完全なソリューション
- 新製品: NGSライブラリー調製キット(推奨)
- V1バックボーンのマウスライブラリーおよびヒトライブラリー:
- Illumina®シーケンスプラットフォーム用Edit-R Mouse Pooled sgRNA Indexing PCR and Sequencing Primer Kit A(必須)
- リバースインデキシングPCRプライマー追加用Edit-R Mouse Pooled sgRNA Indexing PCR and Sequencing Primer Kit B(実験デザインにより追加のマルチプレキシング機能が必要な場合)
重要なお知らせ
Edit-R Lentiviral sgRNAプール化スクリーニング用ライブラリーは、以下に記載されている封じ込め措置および適用される法律および規制が満たされている研究所での内部研究使用のみを目的としています(製品利用規約に規定されているとおり)。製品は、診断、治療、またはその他の商業目的で使用することはできません。また、ヒトに対してはいかなる目的であっても投与することはできず、動物に対しては治療目的で投与することはできません。レンチウイルス粒子として提供されるEdit-R Lentiviral sgRNAプール化スクリーニング用ライブラリーは、複製能力がなく、自己不活化(self-inactivating: SIN)であり、非病原性(感染性ヒト疾患を引き起こさない)です。
レンチウイルス粒子製品を購入する研究者は、レンチウイルスベクター粒子の取り扱いに関する特定のガイドラインについて、所属機関の健康およびバイオセーフティ担当者と相談する責任があります。さらに、各研究者は、複製能のないSINレンチウイルスベクターおよび複製欠損レンチウイルス粒子を使用した研究について、その地域の管轄区域および機関における必要な許可を取得する全責任を負います。
** 詳細については、Scientific supportまでお問い合わせください
それぞれのヒトまたはマウスEdit-R Lentiviral sgRNAプール化ライブラリーにおける標的遺伝子の概数です。各ライブラリーの遺伝子構成に関する詳細については、Scientific Supportまでお問い合わせください。
| Approximate Number of targeted genes | ||
|---|---|---|
| Human | Mouse | |
| Whole Genome | 19,000 | 19,680 |
| Druggable Genome | 8,350 | 9,930 |
| GPCRs | 480 | 510 |
| Ion Channels | 430 | 340 |
| Protein Kinases | 760 | 780 |
| Phosphatases | 310 | 270 |
| Proteases | 700 | 700 |
| Ubiquitin Conjugate | 660 | 660 |
| Epigenetics | 860 | 770 |
| Cytokine Receptors | 140 | 140 |
| Membrane Trafficking | 140 | 110 |
| Deubiquitinating Enzymes | 110 | 100 |
| Cell Cycle Regulation | 170 | 120 |
| Tyrosine Kinases | 90 | 90 |
| Nuclear Receptors | 50 | 50 |
| Apoptosis | 550 | 290 |
| DNA Damage Response | 240 | |
Edit-R Human Whole Genomeライブラリーを用いたスクリーニングに必要なレンチウイルス粒子量
| Fold representation | Replicates | sgRNAs per gene | Volume of lentiviral particles |
|---|---|---|---|
| 200 | 2 | 4 | 0.07 mL |
| 200 | 2 | 8 | 0.13 mL |
| 400 | 2 | 4 | 0.13 mL |
| 400 | 2 | 8 | 0.25 mL |
| NGS Library Prep Kit Contents | Small: 12 Whole Genome Screens | Large: 24 Whole Genome Screens |
|---|---|---|
| NEXTFLEX® PCR Master Mix (green cap) | 3600 μL | 7200 μL |
| Dharmacon HIT Identification Primers (orange cap) | 300 μL | 600 μL |
| NEXTFLEX® PCR II Barcoded Primer Mix | 4 μL | 4 μL |
| Resuspension Buffer | 12 mL | 24 mL |
| Nuclease-free water | 8 mL | 16 mL |
| Cleanup Beads | 7 mL | 15 mL |
Edit-R Lentiviral sgRNAプール化スクリーニング用ライブラリーを使用した遺伝子ノックアウトスクリーニングワークフロー
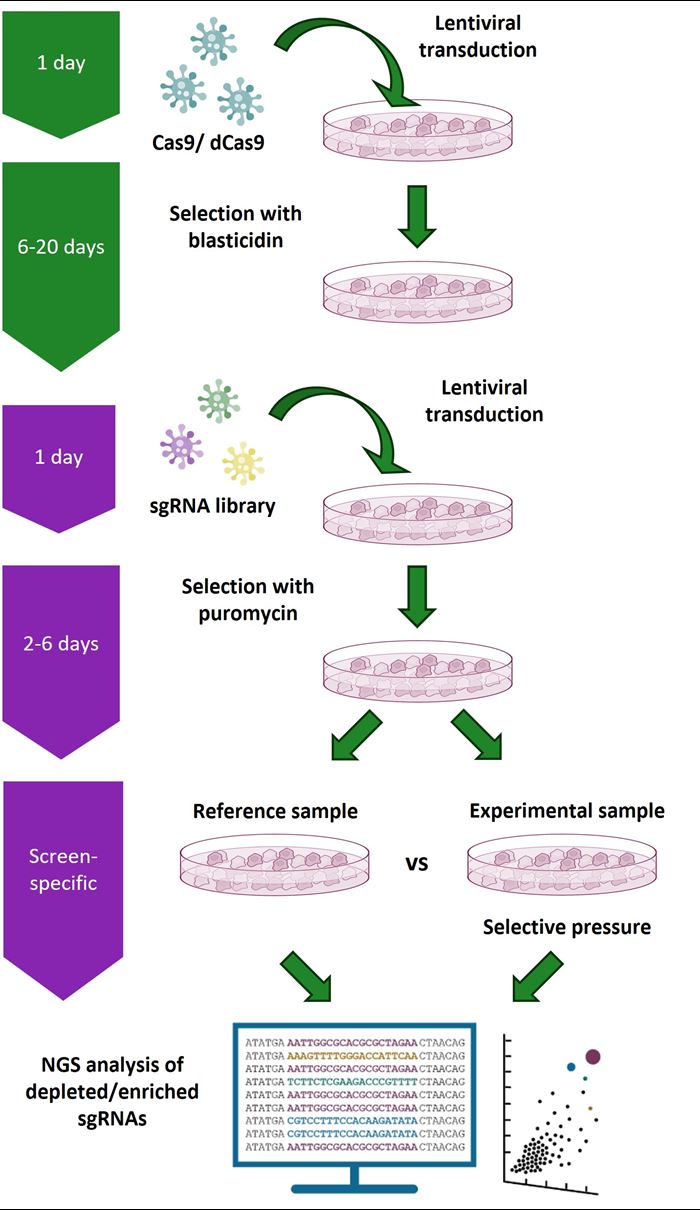
Cas9発現安定細胞株(混合またはクローン細胞集団)は、まずEdit-RレンチウイルスCas9 Nuclease粒子を使用して、ブラストサイジンで選択することにより作製します。次に、これらの細胞にEdit-R lentiviral sgRNAプール化ライブラリーを導入し、ピューロマイシンで選択します。形質導入された細胞は、選択圧および/または表現型選択の適用のためにコントロール集団と実験集団に分割されます。次に、形質導入された細胞のコントロール集団および実験集団からゲノムDNAを抽出し、単離されたgDNA内のsgRNAコンストラクトをベクター特異的なNGSライブラリー調製キットを用いて増幅します。その後、サンプルをマルチプレックス化し、Illumina®プラットフォーム上でシーケンシングが可能です。カスタムシーケンシングリードプライマーは不要です。コントロールサンプルと実験サンプルの両方に組み込まれたsgRNAシーケンスが特定され、相対的な存在量が比較されます。濃縮または枯渇したsgRNAコンストラクトは、追加の表現型および/または生化学的アッセイで個々のEdit-RレンチウイルスsgRNAを使用して確認し、さらに研究するヒットとして同定されます。
Edit-RレンチウイルスCas9ヌクレアーゼおよびsgRNAのベクターマップ
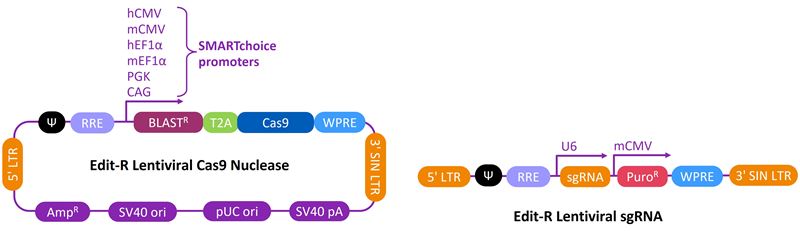
Edit-RレンチウイルスCas9ヌクレアーゼ(A)およびsgRNA(B)ベクターのベクターマップとベクター要素。最大のCas9発現のためのオプションと、目的の標的サイトに合わせて設計されたsgRNA発現のための遺伝子特異的ベクター。
高品質のプール化スクリーニングは、厳密なレンチウイルスsgRNAプール化ライブラリーの作製から始まる
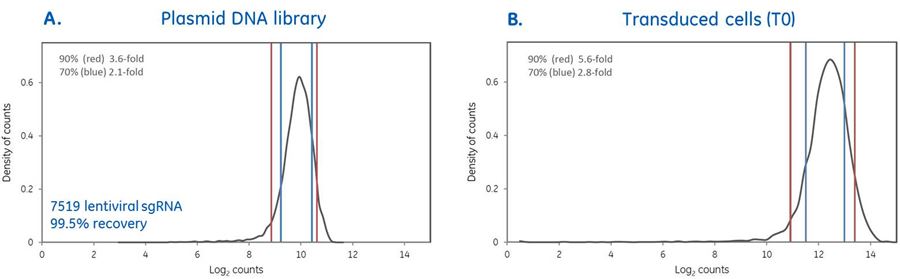
高品質のプール化スクリーニングは、厳密なレンチウイルスsgRNAプール化ライブラリーの作製から始まります。ヒトキナーゼを標的とする7519個のレンチウイルスsgRNAで構成されるプール化ライブラリーを作製し、プラスミドDNAライブラリーの品質を次世代シーケンシング(NGS)によって検証しました。マッピングされたリード100万回あたりのカウントを取得して、入力sgRNAの回収率(99.5%)を決定しました。プール内のsgRNAの70%の存在量が互いに2.1倍の差であり、プール内のsgRNAの90%の存在量が互いに3.6倍の差であることを確認できました(パネルA.)。次に、プールしたプラスミドDNAライブラリーをレンチウイルス粒子にパッケージングし、MOI 0.3でU2OS細胞に形質導入し、形質導入後24時間(T0)にゲノムDNAを調製し、PCR増幅し、NGS用に準備しました。レンチウイルスsgRNAの分布は実質的に影響を受けず、厳密な生産と、生産からプール化スクリーニングへのコンストラクトの保持が最大化されたことを示しています(パネルB.)。
プール化レンチウイルスsgRNAヒトキナーゼライブラリースクリーニングは、信頼性の高い生存率のヒットを同定する
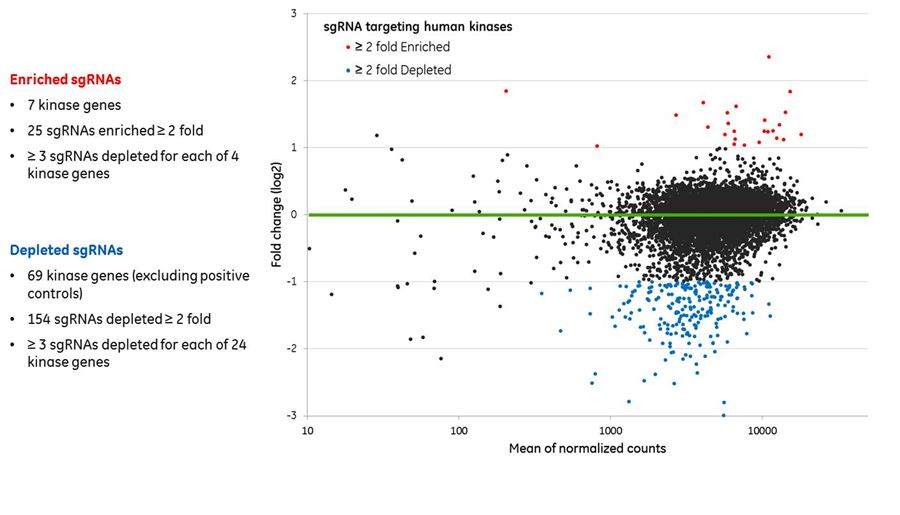
プール化レンチウイルスsgRNAヒトキナーゼライブラリースクリーニングは、信頼性の高い生存率のヒットを同定します。708のヒトキナーゼを標的とする7079のEdit-R sgRNA、34の必須遺伝子を標的とする340のポジティブコントロールsgRNA、および100のnon-targetingコントロールsgRNAで構成されるプール化レンチウイルスライブラリーを、MOI 0.3および1000倍のsgRNA representationでCas9発現U2OS細胞株に形質導入しました。2つの生物学的レプリケートを使用します。形質導入の24時間後に、コントロール細胞集団(T0)を採取し、実験細胞集団(T1)を2 ug/mLピューロマイシン処理(選択圧)にかけました。選択後8日目にT1細胞集団を採取しました。T0およびT1サンプルの両方からゲノムDNAを抽出し、PCR増幅し、Illuminaプラットフォームを用いたハイスループットシーケンシングを行いました。sgRNAの存在量は、正規化されたカウントデータのMAプロットとして表示されます。有意に(adjusted p-value ≤ 0.05)量が多く、量が少なく、2倍以上の量の変化を示したsgRNAは、それぞれ赤と青です。緑色の線は、存在量の変化が0倍であることを示しています。
