Considerations for design of an optimal ssDNA donor for CRISPR-Cas9-mediated precise gene editing
Insertion of an epitope tag, mutation or other genomic modification can be readily accomplished by using CRISPR-Cas9 to cause a double-strand break (DSB) followed by homology-directed repair (HDR) using a repair template (donor oligo or donor plasmid). Knock-in of shorter sequences such as a single nucleotide (nt) change, or insertion of exogenous DNA up to ~100 nts can be performed with a single-stranded, synthetic DNA oligo (Figure 1). The design of a single-stranded donor template plays a big role in your knock-in efficiency; important parameters include homology arm length, homology arm symmetry and chemical modifications. Our top recommendations for each of these, thanks to our great R&D team, are provided here to help you obtain the best results possible for your experiment.
Designing DNA donor oligos for HDR
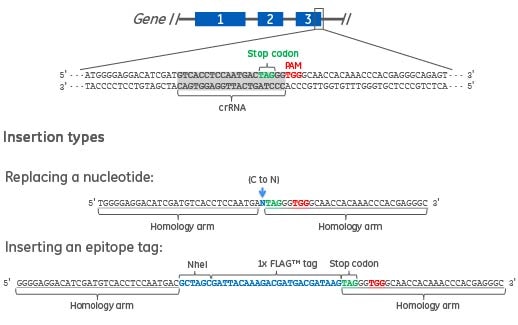
Figure 1. Two types of DNA donor oligos are shown. The first is a 61 nucleotide DNA donor oligo to replace a single nucleotide in the target gene. The homology arms specific to the gene target (underlined) are flanking the nucleotide to be changed (blue text). The second is a 90 nucleotide DNA donor oligo to create a C-terminal epitope tag (e.g. FLAG) and a RFLP detection site (e.g. NheI). The homology arms specific to the gene target (underlined) are flanking the epitope tag and NheI restriction site (blue text).
HDR donor oligo design tips for optimal knock-in efficiency
| Single-stranded DNA donor design parameters | In-house testing results with Edit-R crRNA:tracrRNA and HDR donor designer | Recommendation |
|---|---|---|
| Homology arm length | Homology arm lengths of 30, 40 & 50 nt has optimal knock-in efficiency, while lengths of 60 & 70 nt have lower efficiency. | 30 nt homology arms give optimal knock-in while minimizing the length of the donor template delivered. |
| Homology arm symmetry | For unmodified DNA templates, asymmetric arms showed modest improvement over symmetric arms consistent with the literature (1). However, when phosphorothioated donor templates were used, symmetric arms resulted in the best knock-in efficiency. | Phosphorothioate-modified DNA templates with symmetric homology arms give optimal knock-in. |
| Chemical modifications | For most sequences tested, phosphorothioate-modified DNA templates outperformed unmodified DNA templates. This is consistent with recent publications (2). | Two phosphorothioate modifications on the 5’ and 3’ ends provide a > 2-fold improvement in knock-in efficiency likely due to the improvement in exonuclease resistance in the cell. |
Design a single-stranded DNA oligo donor with our free online tool
Our HDR Donor Designer incorporates these design recommendations (Table 1) based on in-house testing and provides an easy-to-use interface for single-stranded donor template design and ordering.
References
- C. D. Richardson, G. J. Ray, M. A. DeWitt, G. L. Curie, J. E. Corn, Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34, 339-344 (2016).
- J. B. Renaud et al.,Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep 14, 2263-2272 (2016).
Additional Resources
-
Homology-directed repair (HDR) with a DNA donor oligo
Considerations for successful HDR experimental design
-
Edit-R HDR Donor Designer - plasmid
Design and order a plasmid DNA donor kit for insertion of an mKate2 or EGFP fluorescent marker, or a custom insert
-
Homology-directed repair with Dharmacon Edit-R CRISPR-Cas9 reagents and single-strand DNA oligos - Application Note
This article was recently moved from our Featured Articles section on our website to our Blog.

