CRISPRmod CRISPR interference (CRISPRi) allows researchers to down regulate specific gene function by blocking transcription, without editing the DNA. In the Horizon CRISPRi system, a deactivated Cas9 nuclease (dCas9) is fused at the C-terminal end to two transcriptional repressors (SALL1 and SDS3). Visit our CRISPRi applications page to learn more.
CRISPRi dCas9-SAL1-SDS3 is available in mRNA and lentiviral formats.
-
CRISPRi dCas9-SALL1-SDS3 mRNA
Co-transfect or electroporate with CRISPRi synthetic sgRNA, then enrich cell populations with fluorescence or antibiotic resistance selection options. This format is best suited for rapid, transient gene repression studies. -
CRISPRi dCas9-SALL1-SDS3 lentiviral particles
Generate your own stable populations of dCas9-SALL-SDS3 expressing cells, which can then be delivered either synthetic or lentiviral CRISPRi sgRNA. This format is ideal for screening or for extended time point assays.
CRISPRi for gene repression
CRISPR-Cas9 systems are not only for creating genomic double-strand breaks, it can also be used to target promoter regions to inhibit transcription. Endogenous expression of a gene can be specifically and efficiently down regulated using dCas9-SALL1-SDS3 in combination with a gene specific guide RNA in either a synthetic or lentiviral format.
Optimization and enrichment
Enrichment for gene interference can be accomplished using dCas9-SALL1-SDS1 mRNA reagents that co-express either EGFP or PuroR (puromycin resistance) with dCas9-SALL1-SDS3. mRNA can be co-transfected or electroporated along with CRISPRi synthetic guide RNA, enabling visualization of successful delivery and increased knockdown levels.
Promoter options
Promoter activity varies by cell type. Therefore, choosing an optimal promoter for your cells is important for robust gene knockdown. Purified high-titer dCas9-SALL1-SDS3 lentiviral particles are available with three different promoter options (hCMV, mCMV, or hEF1a).
SMARTchoice promoter options for expressing dCas9-SALL1-SDS1| Promoter | Description |
|---|---|
| hCMV | human cytomegalovirus immediate early promoter |
| mCMV | mouse cytomegalovirus immediate early promoter |
| hEF1α | human elongation factor 1 alpha promoter |
CRISPRi dCas9-SALL1-SDS3 mRNA
Purified dCas9-SALL1-SDS3 mRNA for transient expression. Fluorescent and puromycin resistance options available for sorting, enrichment, or visualization of delivery.
CRISPRi dCas9-SALL1-SDS3 lentiviral particles
Purified lentiviral particles for generation of stable dCas9-SALL1-SDS3 expressing cell populations.
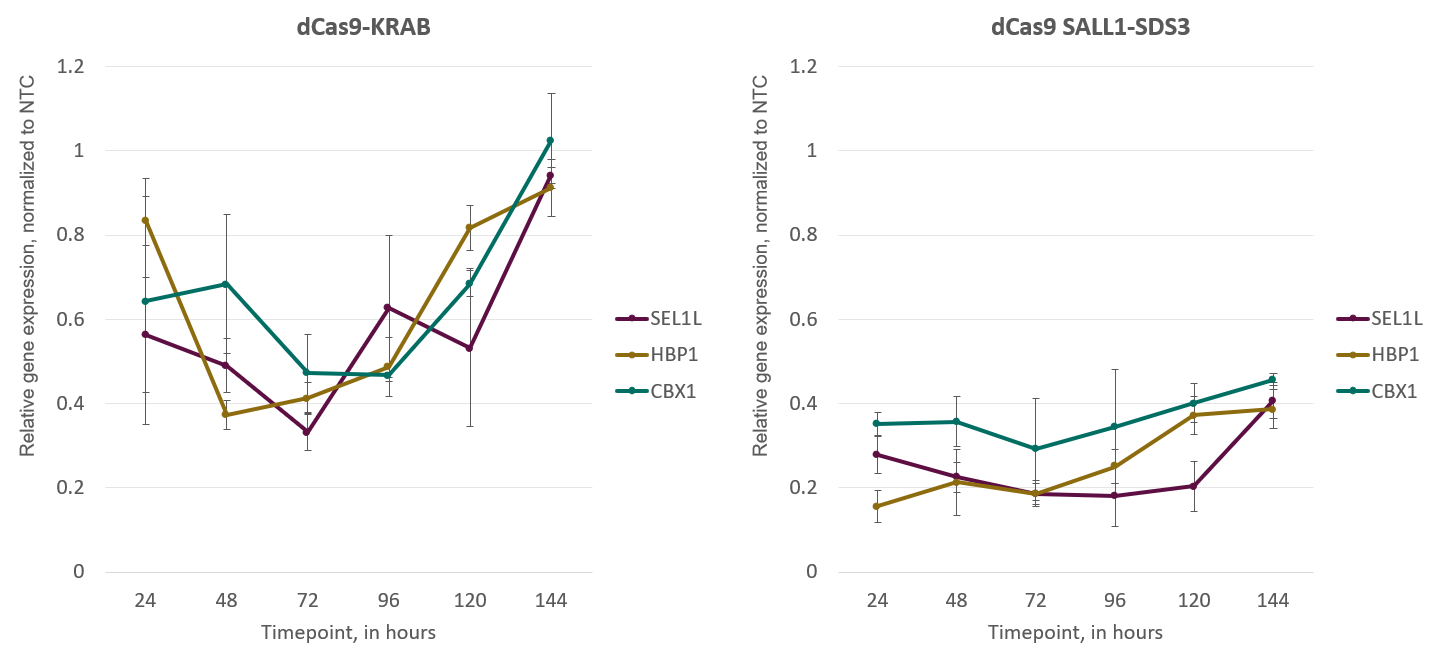
CRISPRi using dCas9-SALL1-SDS1 shows improved gene repression with greater consistency over time when compared to dCas9-KRAB |
 |
|
CRISPRi synthetic sgRNAs achieve maximal repression at 48-72 hours post-transfection in both dCas9-KRAB and Cas9-SALL1-SDS3-expressing cells. U2OS cells stably expressing integrated dCas9-KRAB or dCas9-SALL1-SDS3 were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with pools of pre-designed synthetic sgRNA (25nM) targeting CBX1, HBP1, or SEL1L. Cells were harvested at 24, 48, 72, 96, 120, and 144 hours post-transfection, total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative expression of each target gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC). |
