Edit-R All-in-one lentiviral sgRNA negative controls
バイオインフォマティクスに基づき、ヒトまたはマウスのゲノムのどの遺伝子も標的にしないように設計および検証されたall-in-one non-targetingコンストラクト

CRISPR-Cas9ゲノム編集実験では、sgRNAによって引き起こされる標的ゲノムDNAの編集の生物学的効果と非特異的効果を正確に区別するために、ネガティブコントロールサンプルを使用する必要があります。
Edit-R all-in-one lentiviral sgRNA non-targetingコントロールは:
- CRISPRガイドRNA配列およびCas9が、Edit-R all-in-one遺伝子標的sgRNAと同様に、最適化されたレンチウイルス発現バックボーンにクローン化されています。
- グリセロールストック、または精製された濃縮レンチウイルス粒子(50 µL*、1 x 107 TU/mL以上)をご用意しています。
- ヒトまたはマウスのゲノムのすべての潜在的なPAM隣接標的に対して、少なくとも3つのミスマッチまたはギャップを持つように設計されています。
5種類のネガティブコントロールCRISPR RNA配列が利用可能です。異なる細胞、異なるタイムポイント、異なるアッセイ、および異なる条件に由来する生物学的変動のため、特定の実験の効果的なベースラインを確立するための最良のネガティブコントロールを特定するためには、少なくとも2つのnon-targeting sgRNAをテストすることをお薦めします。
* all-in-one lentiviral sgRNA non-targetingコントロール粒子の大容量をご要望の場合は、お問い合わせください。
Edit-R CRISPR-Cas9 Gene Engineering Platform
The Dharmacon Edit-R Gene Engineering platform is powered by a proprietary algorithm and based on the Type II CRISPR-Cas9 system from the bacteria Streptococcus pyogenes.
- Ready-to-Use Components: The system eliminates the need for cloning, allowing researchers to quickly initiate experiments with predesigned guide RNAs and other essential components, significantly reducing the time to results.
- High Specificity and Efficiency: Edit-R utilizes a proprietary algorithm to design guide RNAs that improve the likelihood of achieving functional knockouts while minimizing off-target effects, ensuring high confidence in experimental outcomes.
- Guaranteed Performance: Every predesigned guide RNA is backed by a guarantee of successful editing at the target site, providing researchers with additional assurance in their gene editing endeavors.
Edit-R デザイン済みガイドRNA保証
すべてのデザイン済みEdit-RガイドRNAは、Edit-Rテクニカルマニュアルの記載に従い細胞導入した場合に、標的領域で編集(DNA切断)ができることを保証します。
Edit-RガイドRNA保証は、野生型S. pyogenes由来Cas9ヌクレアーゼ(タンパク質、mRNA、発現用プラスミド、または発現用レンチウイルスベクター)とともに使用した場合に適用されます。更にEdit-R 化学合成crRNAは、Edit-R tracrRNAとともに使用した場合に適用されます。
T7EIまたはSurveyorミスマッチ検出アッセイを使用して、試薬で処理された細胞集団の編集(DNA切断)の分析結果を提示する必要があります。同時並行で適切に実施されたEdit-R ポジティブコントロールが編集(DNA切断)に成功する一方で、Edit-Rデザイン済みガイドRNAの編集(DNA切断)が成功しない場合、同じフォーマットで同じ容量の、異なるEdit-Rデザイン済みガイドRNAの交換製品が、1回限り無償で提供されます。
交換製品の提供は、テクニカルサポートチームとの話し合いでのみ承認されます。
DNAレベルでの編集(切断)の成功は、常に機能的な遺伝子ノックアウトにつながるとは限りません。複数のガイドRNAをテストして、標的遺伝子のノックアウトに最も効果的なガイドRNAを決定することをお勧めします。
この保証は付随する実験費用には適用されず、CRISPR Design Toolを介して注文されたガイドRNAには適用されず、また本保証により交換したガイドRNAには適用されません。
Edit-R all-in-one lentiviral sgRNAシステムを用いた遺伝子ノックアウトワークフロー

混合集団(左側)またはクローン細胞株(右側)の実験アプローチを用いたall-in-one lentiviral sgRNAによる遺伝子ノックアウトワークフロー
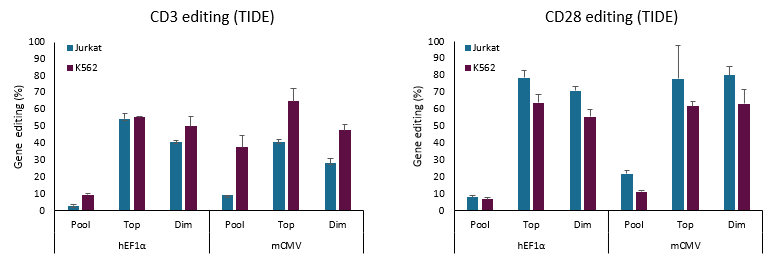
Edit-R all-in-oneレンチウイルスシステムを用いた場合の編集頻度(TIDE)
JurkatまたはK562細胞に、CD3またはCD28遺伝子のいずれかを標的とするEdit-R All-in-one EGFPレンチウイルスsgRNAベクターを用いて、0.3 MOIで形質導入しました。all-in-oneレンチウイルス粒子は、hEF1αまたはmCMVプロモーターの下で構成的にCas9ヌクレアーゼを発現します。Non-targetingコントロール(NTC)ガイドを発現するレンチウイルス粒子をネガティブコントロールとして使用しました。レンチウイルスの形質導入と増殖の後、細胞をソーティングせず細胞集団(Pool)として分析するか、またはソーティングを行って、最も明るい(Top)または最も暗い(Dim)EGFP発現細胞集団を濃縮しました。次に、細胞のPool、Top、およびDimの集団をTIDE分析にかけ、ゲノム編集の割合を評価しました。
Edit-Rの 2ベクターシステムとall-in-oneシステムの比較
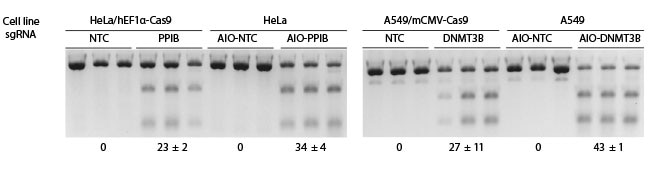
2ベクターシステムの場合、hEF1αまたはmCMVの制御下でCas9を構成的に発現するためのレンチウイルス粒子を細胞に0.3 MOIで形質導入し、10 µg/mLブラストサイジンで10日間選択した後、all-in-one Non-targetingコントロール(NTC)あるいは、DNMT3BまたはPPIBをターゲットとするsgRNAレンチウイルス粒子を0.3 MOIで形質導入しました。all-in-oneシステムでは、all-in-one Non-targetingコントロール(NTC)あるいは、DNMT3BまたはPPIBをターゲットとするall-in-oneレンチウイルスsgRNAを野生型細胞に0.3 MOI にて形質導入し、2 µg/mLピューロマイシンで3日間選択しました。次に、細胞を溶解し、T7EIを用いたDNAミスマッチ検出アッセイを使用してindel(挿入・欠失)を分析しました。
Edit-R all-in-one Lentiviral sgRNAのベクターマップ
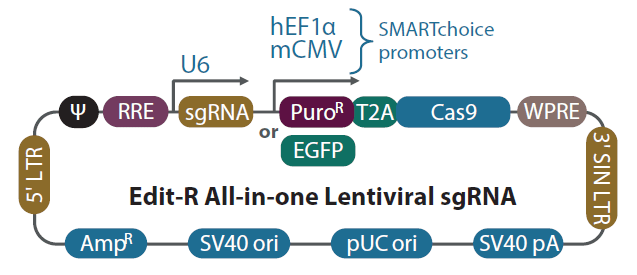
Edit-R all-in-one Lentiviral sgRNAベクターバックボーンでは、遺伝子特異的ガイドRNAはヒトU6プロモーターの制御下で発現し、Cas9およびピューロマイシン耐性マーカー(PuroR)またはEGFPマーカーの発現はヒトEF1αまたはマウスCMVプロモーターのいずれかから駆動されます。プラスミドには、大腸菌での増殖と選択のためのアンピシリン耐性マーカーが含まれています。
| Vector Element | Utility |
|---|---|
| Cas9 | Human codon-optimized S. pyogenes Cas9 nuclease for cleavage of targeted DNA when programmed with a sgRNA |
| T2A | Self-cleaving peptide allows for simultaneous expression of puromycin resistance and Cas9 protein from a single transcript |
| PuroR | Puromycin resistance marker permits antibiotic selection of transduced mammalian cells |
| EGFP | Fluorescent marker permits FACS selection of transduced mammalian cells |
| mCMV | Mouse cytomegalovirus immediate early promoter |
| hEF1α | Human elongation factor 1 alpha short promoter |
| U6 | Human RNA polymerase III promoter U6 |
| sgRNA | Optimized single guide RNA, a fusion of gene-specific crRNA with the tracrRNA scaffold |
| 5' LTR | 5' Long Terminal Repeat necessary for lentiviral particle production and integration of the construct into the host cell genome |
| Ψ | Psi packaging sequence allows lentiviral genome packaging using lentiviral packaging systems |
| RRE | Rev Response Element enhances titer by increasing packaging efficiency of full-length lentiviral genomes |
| WPRE | Woodchuck Hepatitis Post-transcriptional Regulatory Element enhances transgene expression in target cells |
| 3' SIN LTR | 3' Self-inactivating Long Terminal Repeat for generation of replication-incompetent lentiviral particles |
| SV40 pA | Simian virus 40 polyadenylation signal |
| pUC ori | pUC origin of replication |
| SV40 ori | Simian virus 40 origin of replication |
| AmpR | Ampicillin resistance gene for vector propagation in E. coli cultures |
Product inserts
Protocols
Safety data sheets
Related Products
DNA二本鎖切断およびゲノム編集の効率を最大化するための実験条件の検討、および最適化の確認のために使用するコントロール