- Non-mammalian research tools
- Yeast Insertional Mutant Collection
Yeast Insertional Mutant Collection

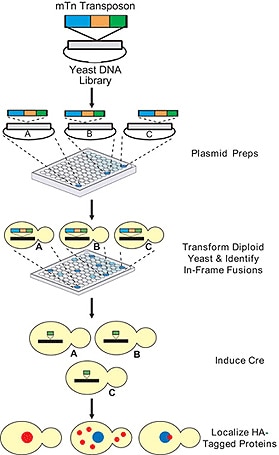
The Yeast Insertional Mutant Collection was developed via transposon insertional mutagenesis. The mini- transposons (mTn) are initially introduced into a plasmid library of yeast genomic DNA, followed by transformation of the disruption alleles into a heterozygous diploid strain of yeast. Once introduced into the yeast, the transposons integrate into the corresponding genomic locus by homologous recombination. The insertion of the transposon into the open reading frame (ORF) of the gene typically disrupts gene function or insertion upstream or downstream of the ORF may result in mis-expression of the gene.
Features
The 6 kb mTn contains the reporter gene lacZ, flanked by lox sites, and a 3xHA tag. The lacZ gene is lacking both its promoter and start codon thus the expression is dependent on the transposon being inserted in-frame into a gene and subsequently transcribed and translated. Using the site-specific DNA recombinase, Cre, the 6 kb insert can be reduced to a 93-codon read-through element containing the 3X HA-tag (see HA-Tagged Yeast Collection).
The mTn technology has been used for large-scale functional analysis of the yeast genome. Access to the resulting gene data, protocols, and publications is available at the Yale Genome Analysis Center website. Information on the insertion site, ORFs affected, and strains available from us can be obtained by searching for the ORF ID on our website
Highlights
- Minitransposons contain the reporter gene lacZ, flanked by lox sites, and a 3xHA tag
- mTn technology can be used for large-scale functional analysis of the yeast genome
- Background strain is Y800 diploid
Note
We provide certain clone resources developed by leading academic laboratories. Many of these resources address the needs of specialized research communities not served by other commercial entities. In order to provide these as a public resource, we depend on the contributing academic laboratories for quality control.
Therefore, these are distributed in the format provided by the contributing institution "as is" with no additional product validation or guarantee. We are not responsible for any errors or performance issues. Additional information can be found in the product manual as well as in associated published articles (if available). Alternatively, the source academic institution can be contacted directly for troubleshooting.
In most cases, the insertion disrupts gene function. Some insertions, however, may not result in gene disruptions. Additionally, individual researchers should confirm that any phenotype observed is actually linked to the indicated insertion.
Individual Clones
Clones are provided as a live culture in a 2 mL tube. Each tube contains the appropriate medium (see product page of collection) supplemented with 15% glycerol. Within 2 to 3 business days of receiving your order, we will ship your clone at room temperature via express overnight delivery. Clones should be stored at -80°C immediately upon receipt.
Bulk Clone Orders
Orders of 50 clones or greater will be provided as live cultures in 96-well microtiter plates. Each well contains the appropriate medium supplemented with 15% glycerol. Bulk orders are shipped on dry ice via express overnight delivery and should be stored at -80°C immediately upon receipt. Please contact our customer service department for a quote and estimated shipping time on bulk orders.
- P. Ross-Macdonald et al., Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 402(6760), 413-418 (25 November 1999).
- A. Kumar et al., High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol. 328, 550-574 (2000).
- A. Kumar et al., TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res. 28(1), 81-84 (1 January 2000).
Related Products
The Yeast HA-Tagged Collection contains mutagenized yeast strains with triple hemagglutinin (HA) epitope tags, useful for immunolocalization, immunoprecipitation, and analysis of binding sites.
Systematic mutations have been engineered into the Yeast Synthetic Histone H3 and H4 mutants in order to elucidate nucleosome functionality.
