Avoid confusion! DNA cleavage and gene editing are not the same.
One of the most common and straightforward ways to estimate the extent of gene editing in a population of cells is with a DNA mismatch detection, such as the T7EI nuclease assay.1 Following a CRISPR-Cas9 gene knockout experiment, a DNA amplicon sample (PCR product) spanning the genomic target site is generated from the genomic DNA of the cell population. This will contain a mixture of unedited (wild type) PCR products and some with various deletions and insertions around the target site. When the wild type and the edited PCR products denature and reanneal, mismatches and regions of single-stranded DNA are formed. Mismatch detection assays work by using an enzyme that detects and cleaves most types of the single-stranded DNA regions (Figure 1). When run out on a gel, the full length and cut products can be visualized and used as an approximation of editing efficiency.
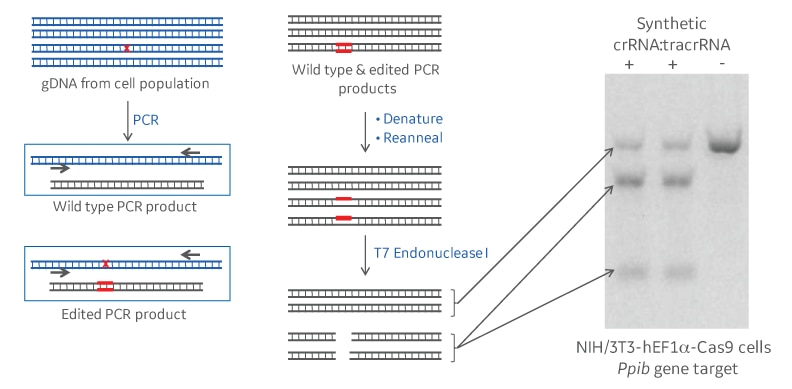
Assessment of efficiency: cleavage vs. editing
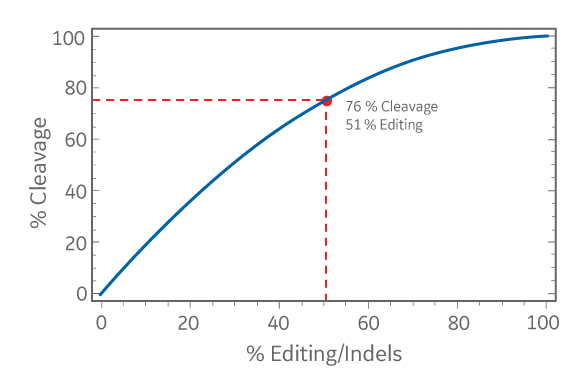
It is important to note that the percent of insertions and deletions in the population is not linearly related to the percent of T7EI cleaved bands. It’s not as simple as a restriction digest!
The percent cleavage and percent editing (insertions and deletions, or indels) are calculated using the following formulas:
% Cleavage = ((a+b)/(a+b+c)) * 100
% Editing/Indels = (1-√(1-(a+b)/(a+b+c)))*100
and the relationship between the two is illustrated graphically; where a and b are intensities of the cut bands in the assay, and c is intensity of the remaining uncut band in the assay.
What this means for CRISPR-Cas9 experimental interpretation
The percent cleavage calculated from the cut bands in the mismatch assay is always an overestimation of the percent editing, or indels. When comparing gene editing efficiency among different samples in one experiment, it’s probably OK to use either calculation method to approximate relative efficiency. However, don’t mix apples and oranges by comparing percent editing to percent cleavage to decide on the best CRISPR reagent or experimental condition between different experiments. A sample with 76 % cleavage will have less editing than a reagent calculated to form 55 % indels (see the graph above). Finally, although the mismatch detection assay is an easy and quick method to estimate relative gene editing, it invariably underestimates efficiency because mismatch detection nucleases are not 100 % efficient at detecting all types of mismatches or bulges/distortions in DNA.2 If you want to accurately determine the editing efficiency and you have a lot of time and resources for that one sample, sequencing is the best method.
Authors: Emily M. Anderson, Senior Scientist and Matthew R. Perkett, Bioinformatics Developer at Dharmacon
References
- R. D. Mashal, J. Koontz, J. Sklar, Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet 9, 177-183 (1995).
- L. Vouillot, A. Thelie, N. Pollet, Comparison of T7EI and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5, 407-415 (2015).
Additional Resources
CRISPR Guide RNA
- Lentiviral and synthetic reagents for targeted gene knockout
Edit-R Synthetic Positive crRNA Controls and Detection Primers
- Species-specific crRNAs targeting well-characterized genes, as well as mismatch detection assay primers, to determine the effectiveness of your gene editing conditions for maximal efficiency.
