Edit-R デザイン済みlentiviral sgRNA(レンチウイルスsgRNA)
効果的かつ正確な遺伝子ノックアウトを実現するシングルガイドRNA発現ベクター
- 目的の標的遺伝子の編集(DNA切断)が保証されています。
- 形質導入に対応したガイドRNAは、クローニングとin vitro転写ステップを必要としません。
- タンパク質の機能的ノックアウトの可能性を最大化し、オフターゲット編集を最小化するようにデザインされています。
- レンチウイルス粒子あるいは大腸菌グリセロールストックをご用意しています。

Edit-R デザイン済みlentiviral sgRNA
1Start Here
2Choose
lentiviral sgRNAは信頼効率的な遺伝子ノックアウトと高い特異性を提供します。
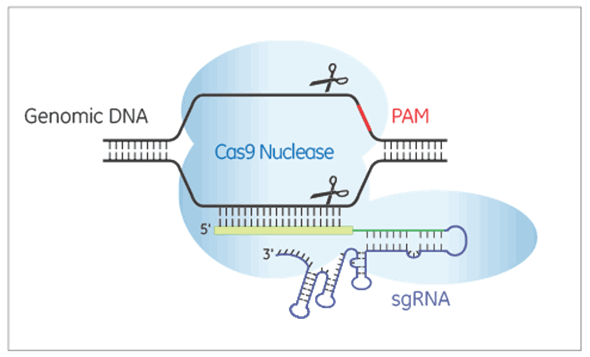
Edit-R lentiviral sgRNAは、Cas9ヌクレアーゼによる標的DNAの二本鎖切断をガイドするためのRNAを発現します。Edit-R lentiviral sgRNAベクターバックボーンでは、遺伝子特異的ガイドRNAはヒトU6プロモーターの制御下で発現されますが、ピューロマイシン耐性マーカー(PuroR)の発現はマウスCMVプロモーターから駆動され、sgRNAが組み込まれた細胞の迅速な選択を可能にします。
各Edit-R lentiviral sgRNAは、目的の遺伝子およびゲノム部位に特異的です。トランスフェクションが困難な細胞でノックアウトを起こす、またはプール化lentiviral sgRNAライブラリースクリーニングによるヒットの更なるフォローアップなどが、このガイドフォーマットの主要な使用方法です。
CRISPR-Cas9は遺伝子機能を調べるための非常に効果的なツールですが、すべてのガイドRNAがタンパク質の機能的なノックアウトを実現するのに有効であるとは限りません。この問題に対処するために、Dharmaconは、挿入または欠失を起こすだけでなく、機能的な遺伝子ノックアウトを生じる可能性が最も高いガイドを選択するように検証されたアルゴリズムを開発しました。
New! Edit-R human sgRNA designs have been updated to the latest RefSeq in 2025 providing the most specific and genomically relevant guides for producing efficient protein knockout. This allows the Edit-R algorithm to target the latest genome annotations more accurately and efficiently providing you with the best solution for your research needs. Please reach out to Scientific Support if you have any questions
すべてのガイドRNAデザインは、各遺伝子についてアルゴリズムで選択した最上位のものです。機能性と特異性の定性的なランク付けにより、特定のアプリケーションに合わせて最適なヒトガイドRNAを選択できます。機能性スコアは、このガイドがどの程度機能的ノックアウトをもたらす可能性があるかを予測したものです。特異性スコアは、潜在的なオフターゲット部位での切断活性の予測リスクに基づいています。詳細については、Edit-RガイドRNAのアルゴリズムページをご覧ください。
Edit-RガイドRNA保証
すべてのデザイン済みEdit-RガイドRNAは、Edit-Rテクニカルマニュアルの記載に従い細胞導入した場合に、標的領域で編集(DNA切断)ができることを保証します。
Edit-RガイドRNA保証は、野生型S. pyogenes由来Cas9ヌクレアーゼ(タンパク質、mRNA、発現用プラスミド、または発現用レンチウイルスベクター)とともに使用した場合に適用されます。更にEdit-R 化学合成crRNAは、Edit-R tracrRNAとともに使用した場合に適用されます。
T7EIまたはSurveyorミスマッチ検出アッセイを使用して、試薬で処理された細胞集団の編集(DNA切断)の分析結果を提示する必要があります。同時並行で適切に実施されたEdit-R ポジティブコントロールが編集(DNA切断)に成功する一方で、Edit-Rデザイン済みガイドRNAによる編集(DNA切断)が成功しない場合、同じフォーマットで同じ容量の、異なるEdit-Rデザイン済みガイドRNAの交換製品が、1回限り無償で提供されます。
交換製品の提供は、テクニカルサポートチームとの話し合いでのみ承認されます。
DNAレベルでの編集(切断)の成功は、常に機能的な遺伝子ノックアウトにつながるとは限りません。複数のガイドRNAをテストして、標的遺伝子のノックアウトに最も効果的なガイドRNAを決定することをお薦めします。
この保証は付随する実験費用には適用されず、CRISPR Design Toolを介して注文されたガイドRNAには適用されず、また本保証により交換したガイドRNAには適用されません。
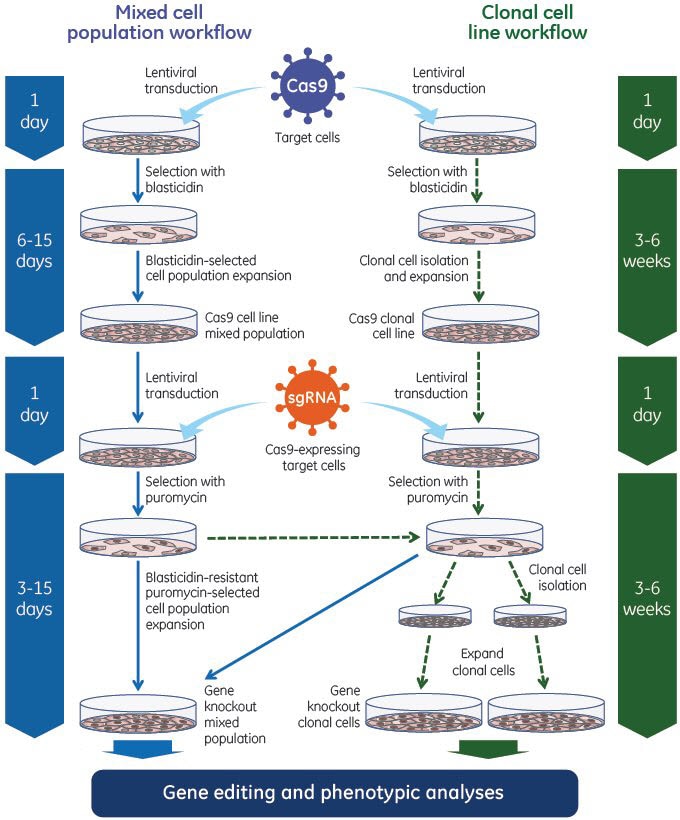
Edit-R lentiviral sgRNAを用いた実験的ワークフロー
Edit-R lentiviral sgRNAを用いたゲノム編集システムでは、Cas9ヌクレアーゼとシングルガイドRNAを2段階のプロセスで利用します。まず、Edit-R lentiviral Cas9ヌクレアーゼ発現粒子を使用し、Cas9ヌクレアーゼを安定的に発現する細胞株を作製します。その後、これらの細胞にEdit-R lentiviral sgRNA粒子を形質導入し、細胞集団または単離されたクローン細胞株の表現型解析のための効率的なゲノム編集(低MOIであっても)を実現できます。
Edit-R lentiviral sgRNAコントロール
-
ポジティブコントロールと検出プライマー
十分に特徴付けられた遺伝子を標的としており、ゲノム編集の効率を最大化するための実験条件の検討、および最適化の確認のために使用します。
-
Non-targetingコントロール
標的遺伝子特異的sgRNAの非存在下でCRISPR-Cas9コンポーネントに対するベースラインの細胞応答を評価するために使用します。
-
Cutting controls
Lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human or mouse genomes.
さまざまなプロモーターの制御下のEdit-R lentiviral Cas9は、複数の細胞株で、Edit-R lentiviral sgRNA用いて高レベルのゲノム編集が実現できることを示す

さまざまなプロモーターの制御下のCas9を用いたゲノム編集テストでは、複数の細胞株で、Edit-R lentiviral sgRNA用いて高レベルのゲノム編集が実現できることが示されています。組み換えU2OSユビキチン-EGFPプロテアソーム細胞株(Ubi [G76V] -EGFP)とHEK293T細胞は、Cas9とブラストサイジン耐性遺伝子を含むレンチウイルス粒子で安定的に形質導入されました。安定的に形質導入された細胞の集団は、sgRNAの形質導入の前に最低10日間ブラストサイジンで選択しました。低MOIでsgRNAレンチウイルス粒子を細胞に形質導入して、細胞当たり1つのsgRNA配列を有する細胞を得て、分析の前に7日間ピューロマイシンで選択しました。ピューロマイシン選択細胞におけるゲノム編集の相対頻度は、T7エンドヌクレアーゼIを使用したDNAミスマッチ検出アッセイから計算しました。
Edit-R lentiviral sgRNAベクターのプラスミドベクターマップ
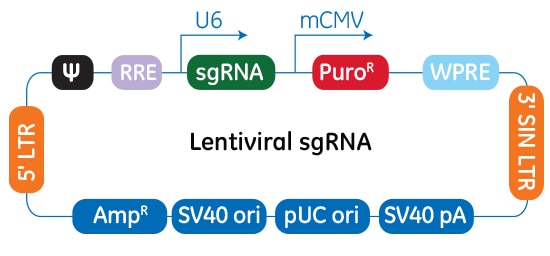
Edit-R lentiviral sgRNAベクターバックボーンでは、遺伝子特異的なsgRNAはヒトU6プロモーターの制御下で発現します。ピューロマイシン耐性マーカー(PuroR)の発現はマウスCMVプロモーターから駆動され、sgRNAが組み込まれた細胞の迅速な選択を可能にします。このプラスミドには、大腸菌での増殖と選択のためのアンピシリン耐性マーカーが含まれています。
アルゴリズムは化学合成crRNAと発現sgRNAの双方に適用
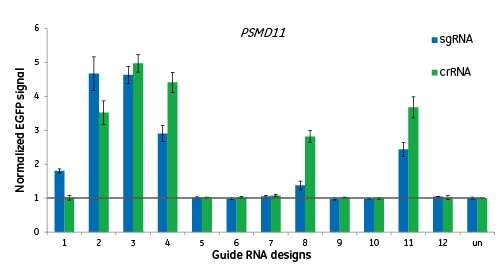
ここでは、12の異なるサイトで遺伝子PSMD11をターゲットにし、プロテアソームの破壊を示すEGFPシグナルによって機能を測定しています。細胞に、crRNA:tracrRNAをトランスフェクトするか、crRNAと同じターゲット配列を含むsgRNAを発現するレンチウイルス粒子を形質導入しました。同じゲノム部位をターゲットとする場合、2つのガイドRNAフォーマットの間に機能の大きな違いはありませんでした。
DharmaFECT kbトランスフェクション試薬を使用したEdit-R lentiviral sgRNAとCas9発現プラスミドの同時導入を利用したCRISPR-Cas9ゲノム編集
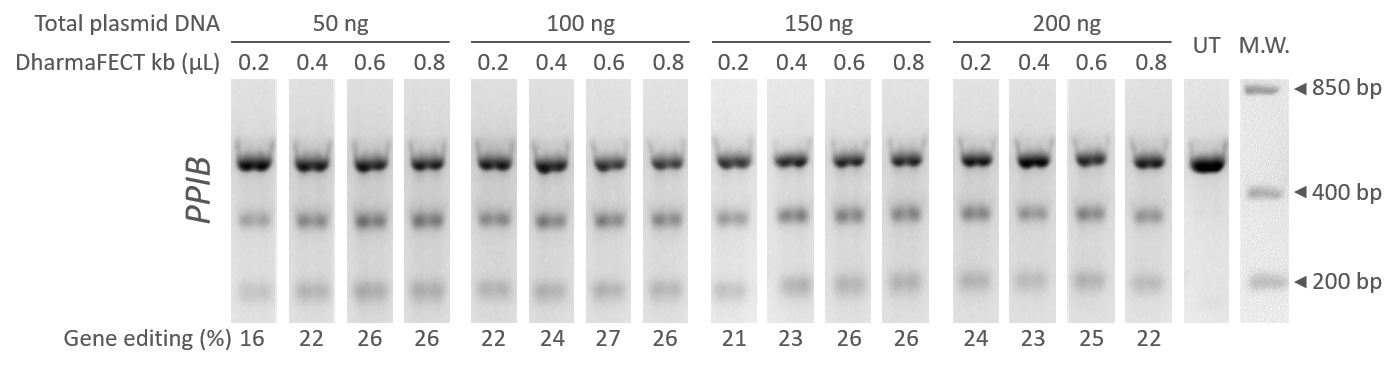
等量のEdit-R hCMV-mKate2-Cas9 Expressionプラスミド(カタログ番号:U-004100-120)およびEdit-R ヒトPPIB lentiviral sgRNAプラスミドを9ウェルフォーマットでU2OS細胞にトランスフェクトしました。トランスフェクションは、総DNA(50~200 ng)およびDharmaFECT kbトランスフェクション試薬(カタログ番号:T-2006-01、ウェルあたり0.2~0.8 µL)の量を何点か変えて行いました。ゲノム編集のパーセンテージは、T7EIを用いたDNAミスマッチ検出アッセイとゲルデンシトメトリーによって、トランスフェクションの72時間後に評価しました。
LentiBOOST Lentivirus Transduction Enhancer is a uniquely formulated transduction reagent that can be used with or without lentivirus spinfection in order to increase successful viral transduction events while preserving cell viability. Especially critical for preserving precious primary cells from patient cohorts, or, for engineering complex animal models; improving transduction efficiency can save time and costs by increasing the success of each editing/transduction step, or, even avoid the loss of irreplaceable samples. Additionally, LentiBOOST technology is already used in the manufacturing of a number of clinical stage therapies providing the opportunity to demonstrate improved workflow applicability to the clinic.
LentiBOOST can be purchased through the Dharmacon Reagents catalog.
To learn more about LentiBOOST technology visit the Revvity LentiBOOST webpage.
Supporting Data
Improved CD8+ T-cell SMARTvector™ shRNA lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
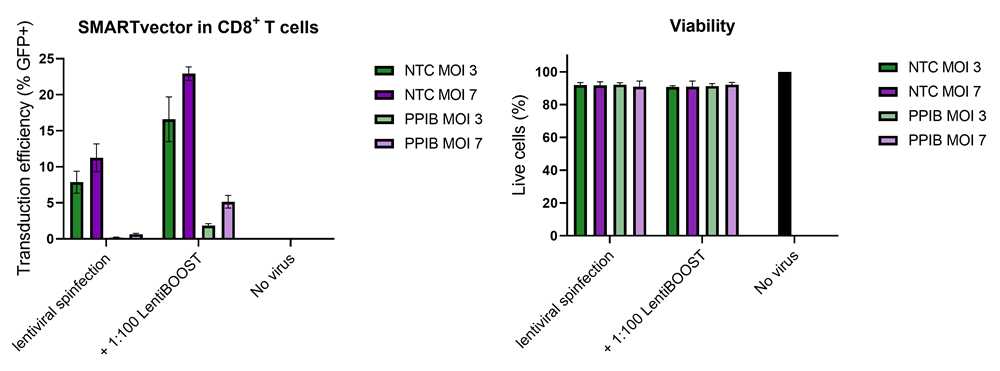
100,000 primary human CD8+ T cells were transduced with either 30,000 (MOI 3, green) or 70,000 (MOI 7, purple) TUs of SMARTVector™ mCMV tGFP Lentiviral Control Particles targeting either NTC or PPIB along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by a four hour incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency (%GFP+ out of live cells) and viability were determined 5 days post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved CD4+ and CD8+ T-cell Edit-R™ All-in-one sgRNA/Cas9 lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
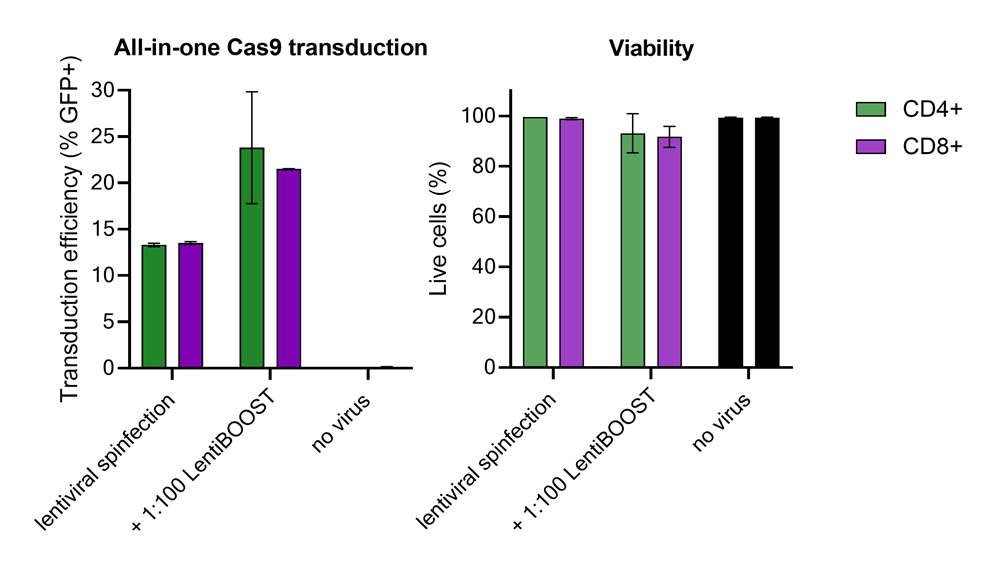
100,000 primary human CD4+ and CD8+ T cells from two donors were transduced with 250,000 TUs of Edit-R GFP Delivery controls mCMV along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved transduction of human induced pluripotent stem cells (hiPSCs) with the Strict-R™ Inducible CRISPRa lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer

10,000 WTC hiPS cells were transduced with either 20,000 (MOI 2, green) or 40,000 (MOI 4, purple) TUs of Strict-R™ Inducible EGFP dCas9-VPR Lentiviral Particles along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST markedly improved transduction efficiencies without significantly impacting cell viability.
Application notes
Posters
Protocols
Safety data sheets
Selection guides
Related Products
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.
バイオインフォマティクスに基づきヒトまたはマウスのゲノムのどの遺伝子も標的にしないように設計および検証されたレンチウイルスsgRNAコンストラクト
DNA二本鎖切断とゲノム編集の効率を最大化するための実験条件の検討、および最適化の確認のために使用するコントロール
