A qPCR primer
Quantitative RT-PCR (qPCR) experiments enable precise and quantitative measurements of gene expression. In the context of mRNA expression experiments, qPCR is often used to quantify changes in relative expression levels between an experimental sample versus a reference sample. Those changes in expression are commonly reported in percent relative to the reference sample, for example to quantify siRNA-mediated mRNA knockdown relative to a non-targeting negative control. (see tech note: Demonstration of a ΔΔCq Calculation Method to Compute Relative Gene Expression from qPCR Data).
In this article, we will focus on the experimental setup, controls as well as validation of results to ensure obtaining reliable mRNA expression data from qPCR experiments.
Setting up your qPCR experiment
Choice of detection chemistry
qPCR relies either on DNA intercalating dyes, or probe-based detection methods to measure amplification of reaction products in real time. Probe-based gene expression assays have the advantage of using a sequence-specific probe to enhance specificity and to deliver reliable results.
Assay design
No matter which detection chemistry you choose, obtaining reliable results depends on knowing your gene, its related transcript variants and SNP locations. PCR primers should be specific and detect all transcripts of interest. In addition, designing exon spanning primers can limit the potential co-amplification of gDNA impurities.
Template quality
Integrity. Sample integrity is critical for obtaining accurate and consistent qPCR results. To inactivate or remove nucleases, workplaces and tools should be regularly decontaminated from potential RNAses. As for any RNA work, gloves should be worn always, and the use of RNAse-free plastics is recommended. Nuclease inhibitors can be used however they will usually only be effective on some subtypes of nucleases. RNA should be stored at -20/-80°C.
Purity. Sample purity is also critical for reliable data. PCR inhibitors should be avoided during all steps of sample preparation. It is of equal importance to make sure no contaminants are present. To prevent gDNA amplification, RNA samples may be treated with either DNAse or Uracil-DNA glycosylase (if dUTP is used for setting up PCR reactions), and primers may be designed to span exon-exon junctions.
Experimental controls and replicates
For obtaining reliable results, it is advisable to include multiple controls and replicates (see figure 1).
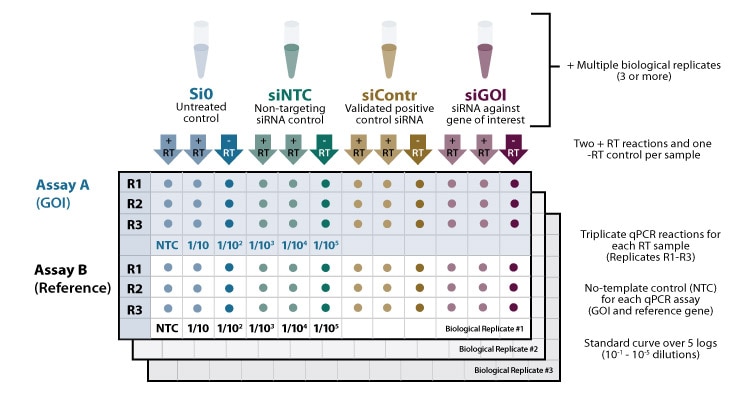
96-well qPCR layout
Replicates
It is recommended to include 3-5 biological replicates for each experimental condition tested to measure experimental variation and determine the statistical significance of results. For each cDNA sample, at least 3 technical replicates are recommended to test assay variability and to minimize pipetting errors.
Controls
Apart from experimental controls (such as untreated, non-targeting and positive controls in the context of RNAi experiments- see Featured Article: Lab Tips - Ensure success with appropriate controls in your RNAi experiments!), the following qPCR controls should be included:
No RT control. For each reverse transcription reaction, a -RT reaction (without reverse transcriptase) is set up to detect gDNA contamination of the extracted RNA that may confuse your mRNA measurement.
No template control. For each gene, a qPCR reaction is set up without cDNA to detect any cross-contamination in the reagents or primers.
Endogenous reference gene. The quantity of mRNA amplified during a qPCR assay will not only depend on gene expression but is also subject to technical variation that may be introduced during RNA extraction and cDNA synthesis, hence normalizing against an endogenous reference gene is recommended.
The expression of the endogenous reference gene should remain unaltered under the conditions and timepoints tested, and be of similar expression level to the experiment’s gene of interest. Testing a large panel of reference genes allows selecting the one that shows the least variation in Cq values (measured as standard deviation) across all tested conditions.
Housekeeping genes such as GAPDH, PPIB, Lamin A/C or ACTB are typically expressed at constant levels, however this is not always the case. Hence, they still need to be tested for variability in expression before using them as endogenous controls.
If there is no single reference gene stably expressed under the conditions tested, using several reference genes in parallel is a well-documented approach.
Standard curve
Standard curves should be performed for any set of qPCR primers as they allow conclusions about the linear range, limit of detection, repeatability and efficiency of an assay. The standard curve is commonly generated using a 10-fold dilution series of at least 5 different standard concentrations in triplicate. All Cq values should fall within the linear range of the standard curve, hence for reliable results, choose Cq values between 15 and 30: Too high target concentrations can have such low Cq values that baseline subtraction becomes difficult- dilution of the sample will enable more accurate measurements. Too low target concentrations that lie below the limit of detection should likewise be excluded from analysis.
Validating your qPCR results
PCR Efficiency
It is recommended to determine the PCR efficiencies for each primer/probe assay. The PCR efficiencies of the target gene and endogenous reference gene should be comparable, for accurate estimation of relative gene expression levels using the ΔΔCq method.
The amplification efficiency will depend on the gene-specific assay, the performance of reaction components, and template quality. To calculate the efficiency of the real-time PCR, the slope of the standard curve can be used. For a PCR efficiency of 100%, the slope of the standard curve would be around -3.3. The correlation coefficient R2 of the curve should be > 0.99 to provide a good confidence.
Limit of detection (LOD)
LOD refers to the minimum quantity of target that can be detected above the background noise of the system. If your gene is expressed at low levels, you may want to check the limit of detection of your assay experimentally, to be sure what you detect is your true target and not just some background noise.
Specificity
In addition to in silico tools like primer BLAST for checking the specificity of PCR primers, a melt curve analysis can be performed to make sure there is only 1 amplification product detected when using SYBR-based primers. Amplification products can be run on an agarose gel to make sure the amplicon size matches the in silico predictions. Probe-based designs use a sequence-specific probe to further enhance specificity.
Summary
qPCR is a powerful technique to study changes in the expression of genes. Appropriate controls and validation experiments should be put in place for each gene being studied. Using the guidelines above will help you achieve robust and reproducible results for your experiments.
Additional Resources

