An overview of design rules, benefits, and limitations of T7EI detection for CRISPR experiments
T7 endonuclease I (T7EI) mismatch detection assays1 are commonly employed to evaluate the gene editing efficiency of CRISPR-Cas9 reagents at a given guide RNA target site in a population of edited cells. Here, we will provide an overview of the method, its advantages and limitations, and describe design considerations for T7EI primers.
Method overview
T7 endonuclease I recognizes and cleaves structural deformities in DNA heteroduplexes. In the context of a gene editing experiment, genomic DNA of a population of cells treated with CRISPR-Cas9 reagents will be amplified by PCR surrounding the CRISPR guide RNA target site. If a non-homologous end joining (NHEJ) repair event following CRISPR-Cas9 cleavage has introduced a mutation, denaturing and annealing will form a fraction of heteroduplexes of mutant and wildtype PCR amplicons. T7 endonuclease I will recognize and cleave DNA mismatches in those heteroduplexes. Running the cleavage products on a polyacrylamide or agarose gel, will resolve full length and cleavage products. The intensity of the respective bands will allow to calculate the gene editing percentage that has occurred (Figure 1).
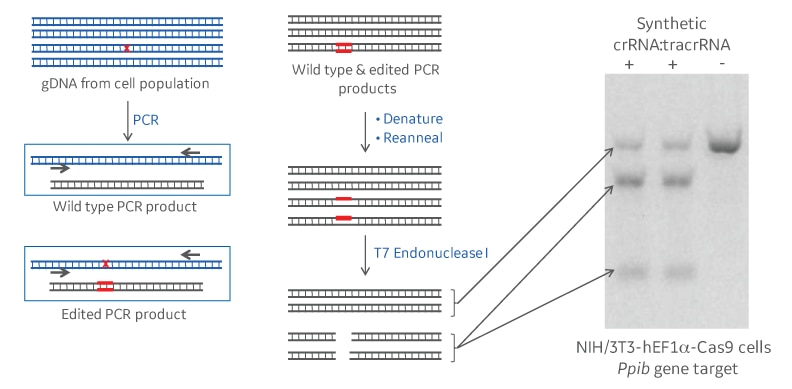
Schematic and example of a DNA mismatch detection assay
Common rules for designing T7 mismatch primers for your gene of interest
PCR primers should surround the desired crRNA cleavage site. We suggest an amplicon size of about 500-800 nucleotides. To be easily resolvable by gel electrophoresis, the cleavage products should be of different sizes (e.g. 150base pairs and 350base pairs), with the CRISPR target site off-center. Primer Tm should match the DNA polymerase that is used for amplification from gDNA.
Utility and limitations
T7EI mismatch detection assays are a quick and cost-efficient means for estimating relative gene editing efficiency in a cell population, or for screening various clones and conditions. They are commonly used to optimize gene editing efficiencies in conjunction with validated guide RNA controls. While they can only assess the relative percentage of insertions and deletions (not functional protein knockout), they are highly useful for:
- Optimizing Cas9/gRNA transfection conditions with a positive control or verifying your experimental conditions for good CRISPR-Cas9 gene editing in ongoing experiments.
- Determining the optimal promoter for driving plasmid-based Cas9 Nuclease expression in your cell line of choice (Figure 2).
- Choosing the most effective guide RNA for knock in experiments by homology-directed repair (HDR), which requires a high cleavage efficiency without the need for a functional knockout.
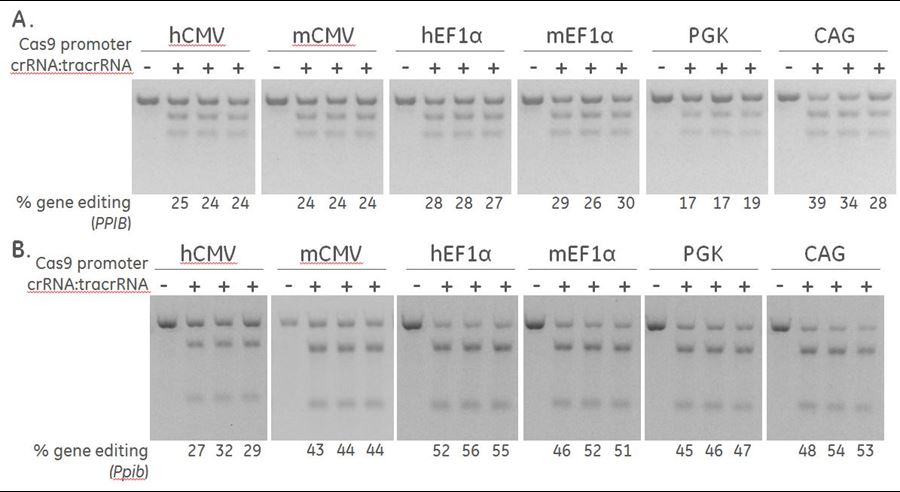
Differential expression of Cas9 by different promoters induce varying levels of gene editing
To assess gene editing by T7EI, keep in mind that DNA cleavage and gene editing are not the same (See featured article: Proper assessment of gene editing with DNA mismatch detection assays). Also, mismatch detection nucleases are not 100% efficient at detecting all types of mismatches or bulges/distortions in DNA2. Likewise, large mutations that mutate the primer binding sites will not be detectable by T7EI mismatch assays.
When using DNA mismatch detection assay to screen mutated clonal lines, T7EI will not be able to recognize homoduplexes of mutant alleles. Clonal lines with the same mutation in all alleles will not be detected as these PCR products can reanneal and not be digested by the endonuclease (See application note: A CRISPR-Cas9 gene engineering workflow: generating functional knockouts using Dharmacon™ Edit-R™ Cas9 and synthetic crRNA and tracrRNA).
Further, the nature of the editing event will not be determined. If you require information on the nucleotide level, sequencing of clonal cell lines is the best method. Sequencing and further functional assays are recommended to fully characterize the impact of the editing event on gene function or cellular phenotype in your edited cell line.
References
- R. D. Mashal, J. Koontz, J. Sklar, Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet 9, 177-183 (1995).
- L. Vouillot, A. Thelie, N. Pollet, Comparison of T7EI and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5, 407-415 (2015).
Author: Kathrin Kerschgens Ph.D. | Project Manager Cell Line Engineering
Additional Resources
CRISPR controls and detection primers
- Ensure your system is optimized for successful CRISPR-Cas9 experiments
The mathematics behind DNA mismatch detection assays
- Learn how to derive the equations behind mismatch detection assays
Beta tool: T7EI calculator
- Automate your T7EI gel analysis to measure percent gene editing using our T7EI calculator!
Proper assessment of gene editing with DNA mismatch detection assays
- This article shows why DNA cleavage and gene editing are not the same and highlights the importance of properly calculating gene editing percentages.
Microinjection of zebrafish embryos using Dharmacon™ Edit-R™ Cas9 Nuclease mRNA, synthetic crRNA, and tracrRNA for genome engineering
- Application example for a T7EI mismatch assay
