- Products
- Dharmaconスクリーニングライブラリー
Human ON-TARGETplus siRNA Library - Epigenetics
siRNA designed and modified for greater specificity

The ON-TARGETplus Human Epigenetics siRNA Library targets enzyme classes characterized to play a role in epigenetic mechanisms (see Reference 1). These inheritable factors that regulate genetic expression are of growing importance to scientific understanding of disease progression and trait inheritance. Modification of histones, such as methylation or acetylation, plays a key role in epigenetics as the extent to which DNA is wrapped around these protein complexes will alter the availability of genes in those DNA segments to be activated. The Epigenetics library targets genes which encode histone and DNA modification factors like methyltransferases, acetyltransferases, kinases and deacetylases, in addition to genes with a role in chromatin remodeling and histone ubiquitination.
The ON-TARGETplus siRNA designs and modifications reduce off-target effects while maintaining high silencing potency for high-confidence screening results.
Highlights
- Patented dual-strand ON-TARGETplus modification pattern on all siRNAs to reduce off-targets
- Sense strand is modified to prevent interaction with RISC and favor antisense strand uptake
- Antisense strand seed region is modified to destabilize off-target activity and enhance target specificity
- Available as SMARTpool siRNA reagents or a Set of 4 siRNAs in 96-well plates
- Guaranteed target gene knockdown (see Specifications tab)
Gene Targets
For a complete list of target genes in this siRNA Library, please contact Scientific Support or your local Sales Representative.
Our siRNA knockdown guarantee
siGENOME and ON-TARGETplus siRNA reagents (SMARTpool and three of four individual siRNAs) are guaranteed to silence target gene expression by at least 75% at the mRNA level when demonstrated to have been used under optimal delivery conditions (confirmed using validated positive control and measured at the mRNA level 24 to 48 hours after transfection using 100 nM siRNA).
Note: Most siGENOME and ON-TARGETplus siRNA products are highly functional at 5 to 25 nM working concentration.
False phenotypes due to off-targets are alleviated by ON-TARGETplus SMARTpool reagents while target gene knockdown is maintained
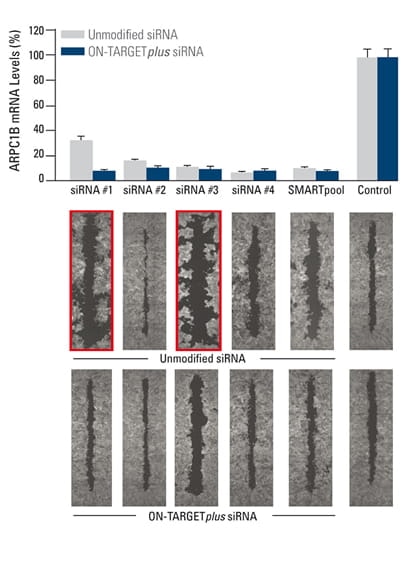
The effect of silencing ARPC1B on cell migration was studied in a breast cancer cell line. A monolayer of cells was uniformly scraped and the rate of cell migration to close the scrape (wound healing) was evaluated. Both unmodified and ON-TARGETplus siRNA reagents induced potent target knockdown. Inconsistent phenotypes due to off-target effects (red outline), were observed for cells transfected with unmodified individual siRNAs. The unmodified SMARTpool improved the false phenotype considerably, while the ON-TARGETplus SMARTpool significantly reduced off-target effects to produce a consistent phenotype. In collaboration with Kaylene Simpson, Laura Selfors, and Joan Brugge, Harvard Medical School.
ON-TARGETplus modifications reduce the overall number of off-targets and pooling reduces them even further
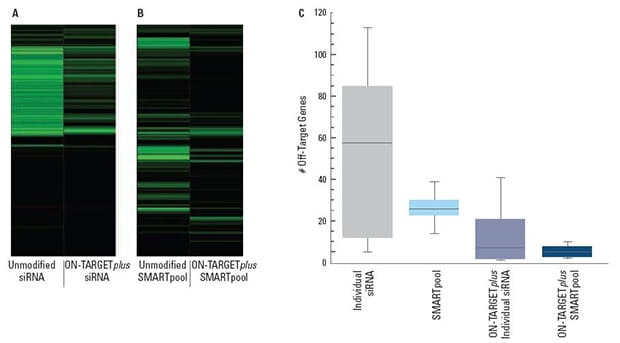
(A) and (B) are representative examples of off-target signatures with and without application of ON-TARGETplus modifications to (A) a single siRNA and (B) a SMARTpool reagent. Green bars indicate genes with 2-fold or more reduction of expression when treated with the indicated siRNA reagent. The ON-TARGETplus modifications reduced the off-targets when compared to unmodified siRNA. Pooling of siRNA and the ON-TARGETplus modification pattern independently, and in combination, provide significant reduction in off-target gene silencing. Panel (C) represents quantitation of off-targets (down-regulated by 2-fold or more) induced by the indicated siRNA reagents targeting 10 different genes (4 siRNAs per gene or a single SMARTpool reagent). Off-targets were quantified using microarray analysis (Agilent) then compiled. Each shaded box represents the middle 50% of the data set. Horizontal line in box: Median value of the data set. Vertical bars: minimum and maximum data values.
- J. Comley. Epigenetics: An emerging target class for drug screening. Drug Discovery World. Spring 2011, 40-55 (2011).
- B.D. Parsons, A. Schindler, D.H. Evans, E. Foley, A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS One. 4(12), e8471 (2009).
- M. Jiang, R. Instrell, B. Saunders, H. Berven, M. Howell, Tales from an academic RNAi screening facility; FAQs. Brief Funct. Genomics. 10(4), 227-237 (2011). [doi: 10.1093/bfgp/elr016]
- J. Borawski, A. Lindeman, F. Buxton, M. Labow, L.A. Gaither, Optimization procedure for small interfering RNA transfection in a 384-well format. J. Biomol. Screen. 12(4), 546-559 (2007).
- T. Ratovitski, E. Chighladze, E. Waldron, R. R. Hirschhorn, C. A. Ross, Cysteine proteases bleomycin hydrolase and cathepsin Z mediate N-terminal proteolysis and toxicity of mutant huntingtin. J. Biol. Chem. 286(14), 12578-12589 (2011). [Human Proteases]
Safety data sheets
Selection guides
Related Products
Concentrated buffer solution recommended for resuspension and long-term storage of any short, double-strand, or single-strand synthetic RNA molecule. Dilute with RNase-free water prior to use.
Catalog ID:B-002000-UB-100
Unit Size:100 mL
$99.00
Molecular grade water for dilution of 5x siRNA Buffer or resuspension of RNA. RNase-free to prevent degradation of RNA reagents and oligonucleotides.
Catalog ID:B-003000-WB-100
Unit Size:100 mL
$32.00
The most broadly applicable DharmaFECT formulation for optimal siRNA or microRNA transfection into a wide range of cell types for successful RNAi experiments
Catalog ID:T-2001-01
Unit Size:0.2 mL
