Revealing the role of E3 ubiquitin ligases in DNA damage repair
One of the diverse new uses for the HAP1 cell line, one that has begun to draw significant attention, is in the field of DNA damage repair. A recent paper from Minoru Takata's group highlights this important application of this relatively new tool.
Published in Molecular Cell in June, 2017, the paper by Inano et al examined an E3 ligase called RFWD3. E3 ubiquitin ligases are known to have a role in DNA damage repair, particularly in homologous recombination (HR). For example, BRCA1, perhaps the most famous gene among the public today, encodes an E3 ligase that is critical for HR. Defects in the BRCA1 protein often lead to breast cancer.
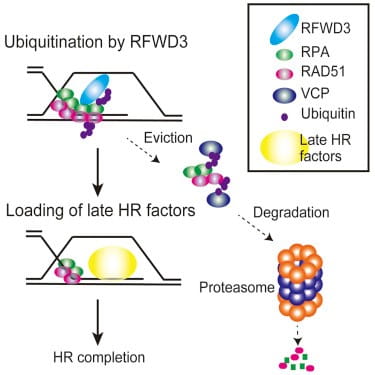
In the paper at hand, the investigators revealed that a primary component of the HR machinery, RAD51, is ubiquitinated by RFWD3. Without functional RFWD3 to ubiquinate them and thereby trigger their removal, RAD51 and another protein RPA (previously known to be an RFWD3 target) remain at the site of DNA repair. This appears to be at least part of the molecular mechanism behind Fanconi anemia, according to another paper published by Takata and colleagues.
According to Takata, If ubiqitination does not occur on these critical HR factors (by RFWD3 knockout, knockdown, or RPA/RAD51 ubiquitination site mutations), [RPA and RAD51] stay at damaged sites and hinder recruitment of downstream proteins. We propose this to be the primary mechanism of HR regulation by RFWD3. The results of deficient RFWD3 functionality include the severe physical challenges faced by FA patients, as well as a dramatically increased risk of cancer.
Like so many molecular pathways, there are multiple related but distinct molecules and processes involved in DNA damage repair. There are numerous E3 ligases known to be involved in triggering removal of repair proteins. The discovery of RFWD3 and subsequent work to delineate its function has the potential to add significant insight into the etiology of a rare and extremely challenging disease in Fanconi Anemia.
For the Takata group, the key to clarifying the role of RFWD3 in DNA damage repair was obtaining a system with complete loss of function of that particular E3 ligase to isolate its specific role from potentially overlapping molecules. The HAP1 cells represented a simple tool to accomplish this. From there, traditional rescue experiments confirmed the specificity of the phenotype and drug sensitivity caused by RFWD3 knockout.
Certainly, an enormous amount of in vitro work then had to be carried out by the group to validate their findings in the HAP1 cells. This was accomplished through an impressive slate of biochemical approaches, as well as traditional knockdown assays through the use of lentiviral siRNA.
Comparisons with other cell models
When asked about how scientists in the field were approaching some of these cell-based assays prior to the use of HAP1 cells, Takata noted that many investigators in the field of DNA damage repair have looked to U2OS cells. For one thing, the U2OS line is marked by a large nucleus, useful for teasing out events taking place on and around the chromosomes. It's worth noting that HAP1 cells, by contrast, have a smaller nucleus. Immunohistochemical approaches are therefore slightly more difficult.
However, U2OS proved problematic: "We tried to knockout RFWD3 in U2OS cells but only got heterozygous KO or in-frame deletions. The possibility to obtain a knockout at (relatively) low cost was very attractive when we [learned of HAP1 cells]."
Thus, the group added HAP1 cells to complement to their arsenal of lab-built CRISPR-based knockout cell lines. The plan is to continue with both HAP1 cells and in-house systems for further investigations into ubiquitin-mediated degradation during HR.
What next?
In addition, the Takata lab is looking to move closer to clinically relevant systems by studying primary cells from humans:
"We are now trying to incorporate iPS cell line into our studies. We are basically looking at a hematological disorder, and the capacity of iPS cells to differentiate in vitro (particularly iPS cells derived from a patient) is very attractive."
Seeing HAP1 cells used in this context is a small indicator of the diverse applications that a well-defined tool can be applied to. However, a far greater value is in studying molecular processes and cellular phenotypes in virtually any context with clean and clear absence of a gene of interest, whether in the nucleus with homologous recombination, or in reverse genetic screens as we will see in a later article.
For more information on DNA damage repair using HAP1 cell lines
Read our blog post on Probing DNA Damage Response Pathways to discover how isogenic cell lines allow single genetic modifications to be studied in isolation.
