Uncovering the truth about the differences between these CRISPR-Cas9 guide RNA formats
Since the advent of CRISPR-Cas9 as a tool for targeted gene knockout, there have been a variety of reagents made available to the research community. Sometimes, understanding the claims about these reagents can be confusing, or offer conflicting data sets in support of a particular product type. In this article we are setting out to do some fact-based myth-busting of common misconceptions regarding two similar CRISPR-Cas9 guide RNA formats: synthetic single guide RNA (sgRNA) and the dual RNA guide of CRISPR RNA (crRNA) and tracrRNA.
Myth 1: Single guide RNA is modeled after “the original” guide RNA
Let’s start with a brief history lesson! The native CRISPR-Cas9 bacterial system was characterized in Streptococcus pyogenes as utilizing a two-part, or dual guide RNA system. Scientists discovered it was a tracrRNA that complexed with a crRNA to recognize and target foreign nucleic acids within bacteria (Figure 1A, Deltcheva 2011). A few years later, the single guide RNA (sgRNA) was created by fusing the crRNA and tracrRNA sequences together into a single RNA chimera by creating a loop at the end of the duplex region (Figure 1B, Jinek 2013, Cong 2013). This allowed for the sgRNA to be expressed from a plasmid or by in vitro transcription for gene editing in mammalian cells.
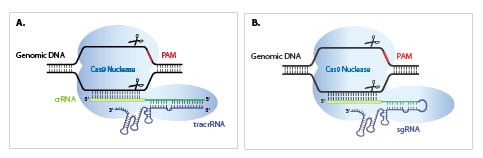
Cas9 nuclease is compatible with both synthetic guide RNA formats
Myth 2: crRNA is more difficult to use than sgRNA
Because crRNA and tracrRNA are separate RNA molecules, it is often assumed that these RNAs need to be heated and cooled to properly anneal them before delivery into cells. In fact, after extensive testing of annealing conditions with Edit-R reagents, we found this is an unnecessary step. Our Edit-R design algorithm considers repeat-antirepeat complementarity in the context of target sequence to ensure good annealing. Simply combining Edit-R crRNA and tracrRNA at room temperature just prior to transfection will result in optimal CRISPR-Cas9 gene editing activity.
Additionally, for arrayed screening libraries of synthetic crRNAs in multi-well plates, the requirement to add tracrRNA may seem like an arduous task. However, these crRNAs can simply be resuspended with Tris buffer that has tracrRNA dissolved in it, or tracrRNA can be incorporated during a dilution step in daughter plate creation. With either method, the number of handling steps is the same as when using sgRNA.
Myth 3: sgRNA gives more efficient gene knockout than crRNA
A look into the performance of the dual RNA system and sgRNA reveals some differences in the levels of editing for individual experiments, but not systemic differences where one format always outperforms the other.
We tested how these guide RNAs compare over a range of concentrations in a Cas9 stably expressing cell line using a well-characterized EGFP reporter system. We targeted genes essential to proteasome function and measured EGFP fluorescence resulting from accumulation of a mutant ubiquitin fused to EGFP. We found that in the case of targeting PSMD7 with either format of guide RNA, similar knockout efficiencies were seen over multiple concentrations. There was a single exception of 6.25 nM where crRNA:tracrRNA outperformed sgRNA by 1.7-fold and a single concentration (50 nM) where sgRNA outperformed crRNA:tracrRNA by 1.3-fold (Figure 2A). This differed when targeting PSMD11, where the dual RNA system functioned better than the sgRNA over three concentrations (Figure 2B).
Our conclusion from these experiments is that neither the sgRNA nor the two-part guide RNA performs universally better than the other, and that differences seen are likely to be due to the cell type and gene target itself.

Knockout performance of synthetic sgRNA and crRNA:tracrRNA
To control for differences in activity between the dual guide RNAs and single guide RNA when Cas9 is not already present within the cell, we tested co-transfection of the guide RNAs with either Cas9 mRNA or Cas9 protein. With co-electroporation experiments in K-562 cells with Cas9 mRNA, we found that when targeting PSMD7, sgRNA produced higher editing efficiency than crRNA:tracrRNA (Figure 3A) but for PSMD11, very similar editing efficiencies were observed with both guide RNAs. Co-electroporation of guide RNAs complexed with Cas9 protein shows there is no difference in editing levels between the dual guide RNA and sgRNA (Figure 3B). Cas9 mRNA co-transfections in U2OS cells produced similar results to co-electroporation where PSMD7-targeting sgRNA produced higher gene knockout than the dual guide RNAs but crRNA:tracrRNA performed similarly to sgRNA for PSMD11 (Figure 3C). Lastly, with Cas9 protein in co-transfections in U2OS, the guide RNA performance was reversed for PSMD7 where crRNA:tracrRNA performed better than sgRNA, similar to what was observed for PSMD11. (Figure 3D).
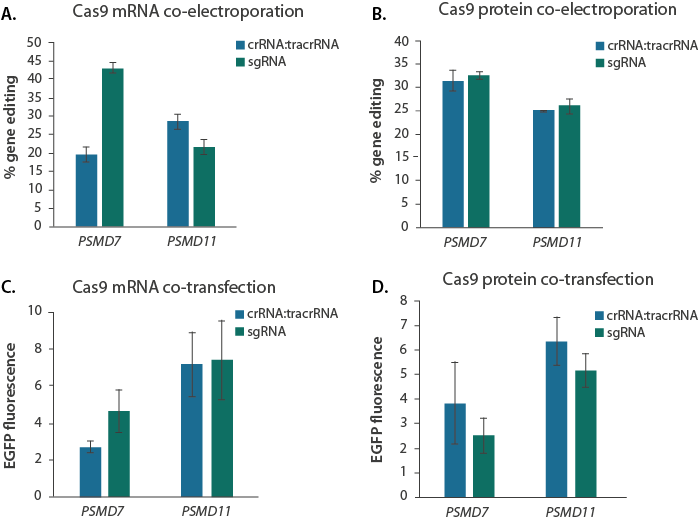
Synthetic guide RNAs co-delivered with Cas9 mRNA or protein
Myths BUSTED
Overall, there will be some differences between the two-part crRNA:tracrRNA and a synthetic sgRNA, but they are minimal, and under most conditions they will function similarly. Discrepancies are most likely to arise from experimental variability from the Cas9 source, delivery method, cell type, or the gene target itself.
We provide both synthetic crRNA:tracrRNA and sgRNA for gene editing and encourage researchers to use the guide RNA reagent that is appropriate for their experimental approach.
Figures 2 and 3 are reformatted from Basila, et al. PLoS ONE 2017.
Author: Megan Basila, Associate Scientist II at Horizon Discovery
References
- E. Deltcheva, et al., CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602-607 (2011) doi.org/10.1038/nature09886
- M. Jinek, et al., RNA-programmed genome editing in human cells. eLIFE 2:e00471 (2013) doi.org/10.7554/eLife.00471
- L. Cong, et al., Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819-823 (2013) 10.1126/science.1231143
- M. Basila, et al., Minimal 2'-O-methyl phosphorothioate linkage modification pattern of synthetic guide RNAs for increased stability and efficient CRISPR-Cas9 gene editing avoiding cellular toxicity, PLoS ONE 12(11): e0188593 (2017) doi.org/10.1371/journal.pone.0188593
Additional Resources
Synthetic crRNA
- Pre-designed or custom synthesized for rapid knockout studies across many genes
Synthetic sgRNA
- Custom single guide RNA oligos for CRISPR-Cas9 gene editing
CRISPR Design Tool
- Place a custom guide RNA order, or design and order your own synthetic sgRNA, crRNA, or lentiviral sgRNA with our easy-to-use interface.
