What you need to know about variables that affect efficiency of HDR knock-in using CRISPR-Cas9
Endogenous labeling of a protein with a fluorescent tag is a powerful technique for understanding native protein function in a living cell through visualization by microscopy (1). Fluorescent-tagging of proteins enables measurement of cell signaling, protein binding, trafficking, function, and kinetics, and is used in a range of applications from in-depth protein mechanism studies to high-throughput screens for drug discovery. The development of CRISPR-Cas9-based tools for HDR (such as Edit-R HDR Donor kits) has dramatically increased the ease of generating a fluorescent protein knock-in.
Insertion of a fluorescent protein into a cell’s genome requires the generation of a double-strand break (DSB) by CRISPR-Cas9, followed by homology-directed repair (HDR) using a repair template such as a plasmid donor. Even with CRISPR-Cas9 technology improvements, knock-in of a fluorescent reporter can cause frustration due to inefficient knock-in rates and a significant workload to screen and identify the clonal cell line that contains the intended genomic modification.
Here are four important considerations that can help you improve the chances of successfully tagging your protein with a fluorescent marker.
-
Fluorescent tagging is most easily applied when the fusion-target protein is robustly expressed and/or localized
-
Use a cell line with high HDR efficiency
-
N- or C-terminal tagging could make your protein non-functional
-
Genomic alterations other than the intended knock-in may impact your results
Protein expression and localization directly affects the feasibility of visualizing a fluorescently tagged protein with a microscope. While a robustly expressed protein is easily visualized, a moderately expressed protein concentrated in a certain location (cellular membrane or organelle) can be more readily observed than one that is diffuse throughout the cytoplasm.
The efficiency of HDR is cell type-dependent and can dramatically affect knock-in efficiency. This was nicely demonstrated in a recent publication (2) where several common cell types’ HDR knock-in efficiencies were compared: K562 cells displayed the highest HDR knock-in efficiency of ~55% while HEK293 cells were much more inefficient at ~ 10%. Choosing a cell type that has a higher HDR efficiency can decrease the workload necessary to isolate the desired clonal cell line and improve the chance of a successful experiment.
Fluorescent-tagging of a target protein fundamentally changes its character and has the potential to alter the protein’s native activity or localization. While fluorescent fusion proteins can give very informative results, systematic studies of N-terminal and C-terminal tagging have shown cases of inactivation, aggregation or non-native localization (3). To reduce wasted time, effort and consumables, it is imperative to examine the literature to determine if tagging your target protein on the N- or C-terminal is more likely to maintain native characteristics. If no information is available, it is worthwhile to create separate fusions, with a fusion at each end, to maximize the chances of success.
Repair of the DSB following CRISPR-Cas9 cleavage is not always exact; mutations such as SNPs, insertions, and/or deletions can occur in one or both alleles in and around the modified genomic site. Even if the expected fluorescent localization of the fusion protein suggests correct function, the fluorescent insert and surrounding sequence must be verified in each knock-in cell line to ensure that fluorescent tagging is the only modification that occurred in the experimental cell line.
A case study: Successful mKate2 tagging of SEC61B in U2OS cells
Protein transport protein Sec61 subunit beta (SEC61B) is a good candidate for fluorescent tagging by HDR using our Edit-R reagents (Figure 1) when we check it against our four considerations:
- SEC61B is known be highly expressed in most cell lines as central component of the protein translocation apparatus of the endoplasmic reticulum (ER) membrane.
- The literature and internal testing have shown good HDR efficiencies in U2OS cells.
- Previous publications have shown that a N-terminal fluorescent protein fusion maintains SEC61B protein function (4).
- Sanger sequencing of our clonal cell line shows homozygous, in-frame insertion of the mKate2 protein after the start codon of SEC61B.
Visualization of mKate2-tagged SEC61B expression
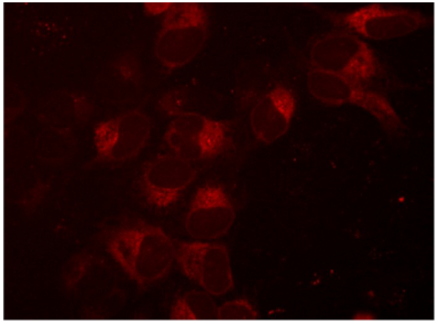
Figure 1. Creation of an mKate2 N-terminal fusion to the SEC61B gene, a component of protein transport in the endoplasmic reticulum. U2OS cells were transfected with 50 nM SEC61B-crRNA:tracrRNA, 25 nM Cas9 protein, 200 ng/µL SEC61B mKate2 HDR donor plasmid, and 0.3 µL/well DharmaFECT Duo transfection reagent and visualized on a fluorescent microscope seven days post-transfection
Learn more about the keys to HDR experimental success »
References
- E. Snapp, Design and use of fluorescent fusion proteins in cell biology. Curr Protoc Cell Biol Chapter 21, Unit 21 24 (2005).
- F. Chen et al., High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8, 753-755 (2011).
- C. Stadler et al., Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods 10, 315-323 (2013).
- Y. Shibata et al., The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283, 18892-18904 (2008).

