ON-TARGETplus 2.0 Cyclophilin B Control siRNA

ON-TARGETplus 2.0 Cyclophilin B Control siRNA is a validated positive control, guaranteed to silence cyclophilin B in human, mouse, or rat cells. The ON-TARGETplus 2.0 siRNA patented dual-strand modification pattern reduces off-targets by up to 90%, allowing improved specificity.
Also known as peptidylprolyl isomerase B, cyclophilin B is associated with the secretory pathway. This gene is abundantly expressed in most cells and because it is non-essential, knockdown of the corresponding mRNA does not affect cell viability.
These controls are recommended for any siRNA workflow utilizing ON-TARGETplus 2.0 reagents.
Highlights
- Validated positive control siRNAs for guaranteed silencing of Cyclophilin B in human, mouse, or rat cells
- Reduced off-targets due to the ON-TARGETplus dual-strand modifications
- Target accession numbers NM_000942 (human, D-001820-01), NM_011149 (mouse, D-001820-02), or NM_022536 or (rat, D-001820-03).
ON-TARGETplus 2.0 maintains high performance of target gene knockdown set by ON-TARGETplus

U2OS or HCT-116 cells were transfected with 25 nM ON-TARGETplus or ON-TARGETplus 2.0 siRNAs targeting the AHCY (left) or PRDX1 (right) genes, or a non-targeting control (NTC) siRNA with 0.2 µL/well DharmaFECT 4 (U2OS) or DharmaFECT 2 (HCT-116). After 48 hours, total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene knockdown was observed with both ON-TARGETplus and ON-TARGETplus 2.0 siRNAs. n = 2 biologically independent samples per group.
ON-TARGETplus 2.0 mitigates newly discovered off-target effects arising from transcriptome expansion

U2OS or HEK293T cells were transfected with 25 nM ON-TARGETplus or ON-TARGETplus 2.0 siRNAs targeting the YY1AP1 gene, or a non-targeting control (NTC) siRNA with 0.2 µL/well DharmaFECT 1 (HEK293T) or DharmaFECT 4 (U2OS). After 48 hours, total RNA was isolated and prepped for RNAseq or RT-qPCR. A. Data plotted as a histogram showing the distribution of global transcriptional expression in U2OS cells, represented as transcripts per million (TPM). Strong downregulation of YY1AP1 mRNA expression was observed in all four siRNA treatment groups, denoted on the x-axis and represented as log2 transformed TPM fold change while global gene expression was largely unchanged relative to the corresponding NTC siRNA. Notably, ON-TARGETplus siRNAs also depleted expression of the off-target gene GON4L whereas ON-TARGETplus 2.0 siRNAs maintained high specificity and did not affect GON4L expression. n = 2 biologically independent samples per group. B. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene knockdown was observed with both ON-TARGETplus and ON-TARGETplus 2.0 siRNAs, however the ON-TARGETplus siRNA was found to also reduce expression of an off-target gene, GON4L. The ON-TARGETplus 2.0 siRNAs maintained high specificity and did not affect GON4L expression. n = 2 biologically independent samples per group.
ON-TARGETplus modifications reduce the overall number of off-targets and pooling reduces them even further

Panels (A) and (B) are representative examples of off-target signatures with and without application of ON-TARGETplus modifications to (A) a single siRNA and (B) a SMARTpool reagent. Green bars indicate genes with 2-fold or more reduction of expression when treated with the indicated siRNA reagent.The ON-TARGETplus modifications reduced the off-targets when compared to unmodified siRNA. Pooling of siRNA and the ON-TARGETplus modification pattern independently, and in combination, provide significant reduction in off-target gene silencing. Panel (C) represents quantitation of off-targets (down-regulated by 2-fold or more) induced by the indicated siRNA reagents targeting 10 different genes (4 siRNAs per gene or a single SMARTpool reagent). Off-targets were quantified using microarray analysis (Agilent) then compiled. Each shaded box represents the middle 50% of the data set. Horizontal line in box: Median value of the data set. Vertical bars: minimum and maximum data values.
Only the ON-TARGETplus modification pattern addresses both siRNA strands for premium specificity
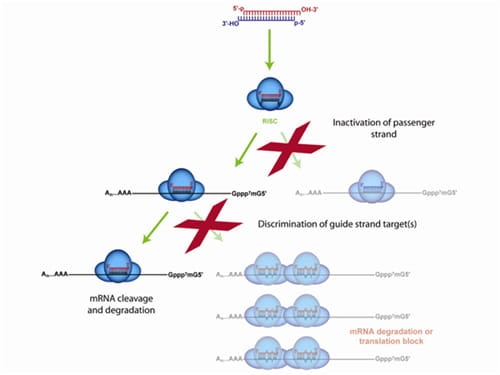
The ON-TARGETplus dual-strand chemical modification begins with the sense (passenger) strand being blocked from RISC uptake to favor antisense (guide) strand loading and reduce passenger strand-induced off-targets. However, the majority of siRNA off-targets are driven by the seed region of the guide strand. ON-TARGETplus is modified within its seed region to destabilize miRNA-like activity and improve specificity to the desired target for potent knockdown.
Protocols
Safety data sheets
Related Products
With ON-TARGETplus 2.0 siRNA, your gene-silencing reagents stay aligned with the most current transcriptome data. Our continuous realignment process ensures every siRNA reflects the latest annotations and delivers consistent, high-performance results.
The most broadly applicable DharmaFECT formulation for optimal siRNA or microRNA transfection into a wide range of cell types for successful RNAi experiments
Catalog ID:T-2001-01
Unit Size:0.2 mL
$116.00
Concentrated buffer solution recommended for resuspension and long-term storage of any short, double-strand, or single-strand synthetic RNA molecule. Dilute with RNase-free water prior to use.
Catalog ID:B-002000-UB-100
Unit Size:100 mL
$99.00
Molecular grade water for dilution of 5x siRNA Buffer or resuspension of RNA. RNase-free to prevent degradation of RNA reagents and oligonucleotides.
Catalog ID:B-003000-WB-100
Unit Size:100 mL
