- 遺伝子発現調節試薬
- RNA干渉
- Dharmacon™ siRNA solutions
- ON-TARGETplus 2.0 siRNA
ON-TARGETplus 2.0 siRNA
Always in Step with the Evolving Transcriptome
With ON-TARGETplus 2.0 siRNA, your gene-silencing reagents stay aligned with the most current transcriptome data. Our continuous realignment process ensures every siRNA reflects the latest annotations and delivers consistent, high-performance results.
What this means for your research:
- Precise Targeting: Designs are routinely reannotated against updated transcriptome databases.
- Enhanced Performance: Outdated sequences are replaced with newly ranked, high-performing designs.
- Reliable Outcomes: Each reagent is optimized for specificity and silencing efficiency.
Stay ahead of evolving biology with siRNA tools that evolve with it.
1Start Here
2Choose
Knockdown your target gene function with confidence
The next-generation RNAi SMARTselection algorithm and design curation for efficient, guaranteed target gene silencing makes ON-TARGETplus 2.0 siRNA the optimal choice for highly effective knockdown with minimal off-target activity.
- Available genome-wide for human, mouse, and rat genes
- Reduced off-targets: next-generation design minimizes unintended interactions compared to unmodified siRNA
- Guaranteed silencing: by SMARTpool and 3 of 4 individual siRNAs (see Guarantee tab)
- Sequence information provided with every siRNA purchase
ON-TARGETplus technology:
Dual-strand strategy: Proprietary synthetic modifications to both siRNA strands. The modified sense strand ensures antisense strand uptake by the RISC; antisense seed regions are modified to reduce off-target activity.
- Seed-region analysis: Filters eliminate common seed sequences likely to cause miRNA-like off-targets
- Seed frequency assessment: Designs minimize sequences with high off-target potential
- Continuous transcriptome alignment: siRNAs are reannotated and ranked to reflect the latest transcriptome data for improved specificity and performance. siRNAs are then curated so that individual siRNAs and SMARTpools of siRNAs maximize coverage of a genes transcript isoforms
With ON-TARGETplus 2.0, your gene-silencing tools are built on landmark studies of seed-region–mediated off-targets, refined with next-generation algorithms for reliable, high-specificity knockdown and consistent experimental outcomes.
ON-TARGETplus 2.0 Key Differentiators
High Specificity - High specificity and minimized off-target effects with advanced SMARTselection design algorithm and proprietary synthetic modification patterns
SMARTpools -Pools of 4 siRNAs further maximize on-target activity and minimize off-target effects while maximizing transcript isoform coverage
Transcriptome-Aligned - Continuously updated to reflect the latest human transcriptome for accurate targeting.
Proven Reliability - Backed by ~30 years of RNA synthesis expertise and validated across thousands of studies
Comprehensive Portfolio - Industry’s largest collection of CRISPR, RNAi, gene editing, modulation, and overexpression tools
Research-Ready Libraries - Genome-wide and pathway-focused collections for complete control over gene function studies
Thought Leadership - Pioneers in RNAi with widely cited algorithm and design publications
Expert Support - Industry-best technical, bioinformatics, and customer service with rapid resolution
Deep Experimental Insight - Innate understanding of research workflows through services, applications labs, and global collaborations
Product formats
- SMARTpool: A mixture of 4 siRNA provided as a single reagent; providing advantages in both potency and specificity.
- Set of 4: A convenient option for purchasing aliquots of all 4 individual siRNAs targeting a single gene.
- Individual siRNAs: For the most precise knockdown of specific transcript isoforms
Components for an RNAi experiment using synthetic siRNA
- ON-TARGETplus 2.0 siRNA
- Appropriate positive and non-targeting controls
- DharmaFECT transfection reagent or electroporation for delivery
Note: You can order guide RNA above, with the option to add any of these additional components directly to your cart.
Order quantity guidelines
ON-TARGETplus 2.0 reagents are routinely used at 5 to 25 nM concentration. The calculations below, based on 25 nM, are for estimation purposes only and assume no loss from pipetting.
| Approximate # reactions (wells) at 25 nM siRNA concentration | |||
|---|---|---|---|
| nmol | 96-well plate (100 µL total reaction volume) | 24-well plate (500 µL total reaction volume) | 12-well plate (1000 µL total reaction volume) |
| 1 | 400 | 80 | 40 |
| 2 | 800 | 160 | 80 |
| 5 | 2000 | 400 | 200 |
| 10 | 4000 | 800 | 400 |
| 20 | 8000 | 1600 | 800 |
Custom siRNA design
Design and order custom siRNA sequences using our siDESIGN Center, available in both legacy ON-TARGETplus and the new ON-TARGETplus 2.0 experience.
Our siRNA knockdown guarantee
ON-TARGETplus 2.0 siRNA reagents (SMARTpool and three of four individual siRNAs) are guaranteed to silence target gene expression by at least 75% at the mRNA level when demonstrated to have been used under optimal delivery conditions (confirmed using validated positive control and measured at the mRNA level 24 to 48 hours after transfection using 25 nM siRNA).
Note: Most ON-TARGETplus 2.0 siRNA products are highly functional at 5 to 25 nM working concentration. For maximizing knockdown we recommend using 100 nM.
Complete your knockout experiment by adding these supporting reagents to your order
ON-TARGETplus 2.0 siRNA controls
Validated positive controls and non-targeting negative controls utilizing patented modifications for optimal specificity. Available as pools or single siRNAs. On-TARGETplus 2.0 control siRNAs and pools are produced with the patented ON-TARGETplus modification pattern for optimal specificity. They are recommended as controls for any siRNA experiment due to their minimized off-target effects.
Our panel of non-targeting controls permits assessment of potential non-specific effects, to find the optimal negative control for your cell type, and your assay. Pooled siRNA controls are recommended for further reduction of off-targets, and for use with SMARTpool siRNA reagents.
Select a species-specific positive control to silence an expressed housekeeping gene in your experimental cell type.
| ON-TARGETplus 2.0 Positive Control Reagents | Species | Catalog Number |
|---|---|---|
| ON-TARGETplus 2.0 Cyclophilin B Control siRNA | Human | D2-001820-01 |
| Mouse | D2-001820-02 | |
| Rat | D2-001820-03 | |
| ON-TARGETplus 2.0 Cyclophilin B Control Pool | Human | D2-001820-10 |
| Mouse | D2-001820-20 | |
| Rat | D2-001820-30 | |
| ON-TARGETplus 2.0 GAPD Control siRNA | Human | D2-001830-01 |
| Mouse | D2-001830-02 | |
| Rat | D2-001830-03 | |
| ON-TARGETplus GAPD Control Pool | Human | D2-001830-10 |
| Mouse | D2-001830-20 | |
| Rat | D2-001830-30 | |
| ON-TARGETplus 2.0 Negative Control Reagents | Species | Catalog Number |
| ON-TARGETplus 2.0 Non-targeting siRNAs | Human, Mouse, Rat | D2-001810-0X |
| ON-TARGETplus 2.0 Non-targeting Pool | Human, Mouse, Rat | D2-001810-10 |
Transfection reagents
DharmaFECT transfection reagents are optimized for improved delivery and reduced toxicity. The optimal DharmaFECT reagent for your experiment will depend on your Cas9 nuclease source and your cell type.
Order quantity guidelines
ON-TARGETplus reagents are routinely used at 5 to 25 nM concentration. The calculations below, based on 25 nM, are for estimation purposes only and assume no loss from pipetting.
| Approximate # reactions (wells) at 25nM siRNA concentration | |||
|---|---|---|---|
| nmol | 96-well plate (100 µL total reaction volume) | 24-well plate (500 µL total reaction volume) | 12-well plate (1000 µL total reaction volume) |
| 1 | 400 | 80 | 40 |
| 2 | 800 | 160 | 80 |
| 5 | 2000 | 400 | 200 |
| 10 | 4000 | 800 | 400 |
| 20 | 8000 | 1600 | 800 |
ON-TARGETplus modifications reduce the overall number of off-targets and pooling reduces them even further
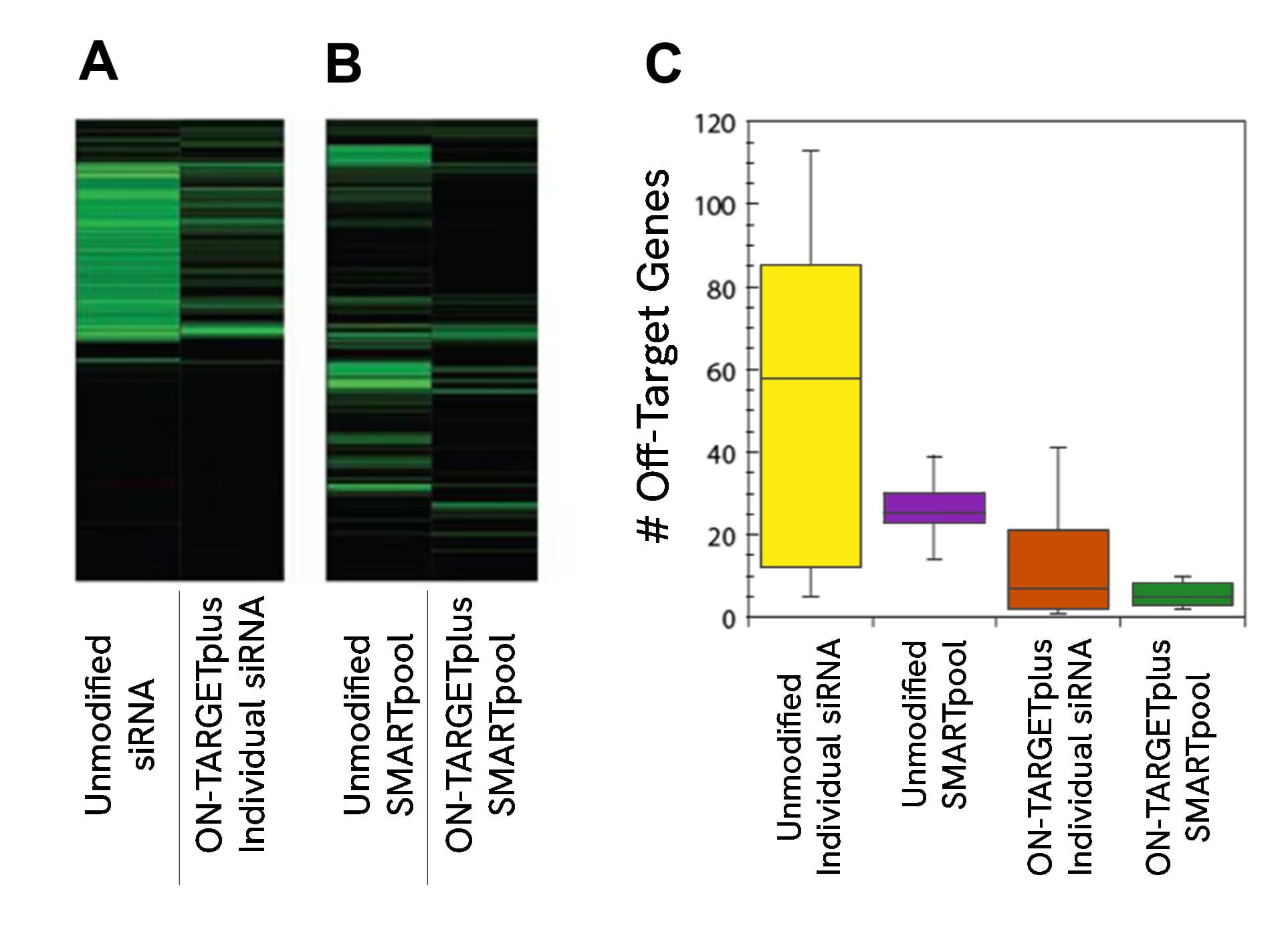
Panels (A) and (B) are representative examples of off-target signatures with and without application of ON-TARGETplus modifications to (A) a single siRNA and (B) a SMARTpool reagent. Green bars indicate genes with 2-fold or more reduction of expression when treated with the indicated siRNA reagent.The ON-TARGETplus modifications reduced the off-targets when compared to unmodified siRNA. Pooling of siRNA and the ON-TARGETplus modification pattern independently, and in combination, provide significant reduction in off-target gene silencing. Panel (C) represents quantitation of off-targets (down-regulated by 2-fold or more) induced by the indicated siRNA reagents targeting 10 different genes (4 siRNAs per gene or a single SMARTpool reagent). Off-targets were quantified using microarray analysis (Agilent) then compiled. Each shaded box represents the middle 50% of the data set. Horizontal line in box: Median value of the data set. Vertical bars: minimum and maximum data values.
False phenotypes due to off-targets are alleviated by ON-TARGETplus SMARTpool reagents while target gene knockdown is maintained
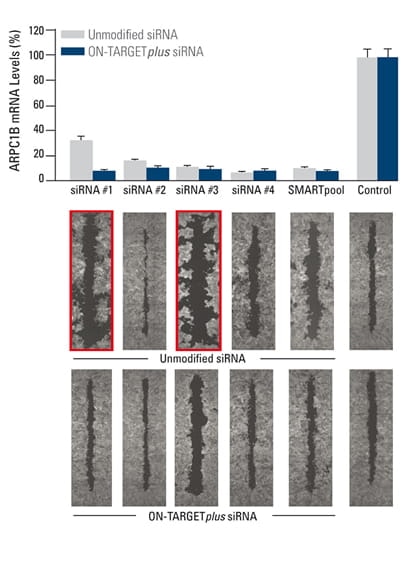
The effect of silencing ARPC1B on cell migration was studied in a breast cancer cell line. A monolayer of cells was uniformly scraped and the rate of cell migration to close the scrape (wound healing) was evaluated. Both unmodified and ON-TARGETplus siRNA reagents induced potent target knockdown. Inconsistent phenotypes due to off-target effects (red outline), were observed for cells transfected with unmodified individual siRNAs. The unmodified SMARTpool improved the false phenotype considerably, while the ON-TARGETplus SMARTpool significantly reduced off-target effects to produce a consistent phenotype.
In collaboration with Kaylene Simpson, Laura Selfors, and Joan Brugge, Harvard Medical School.
Only the ON-TARGETplus modification pattern addresses both siRNA strands for premium silencing
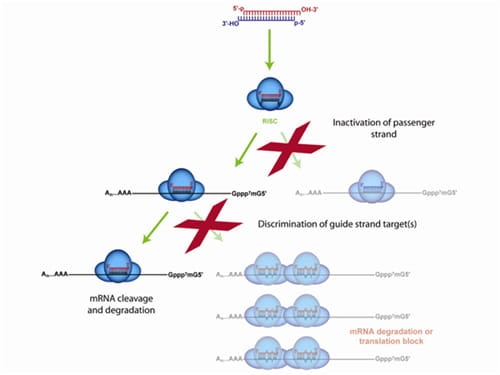
The ON-TARGETplus dual-strand chemical modification begins with the sense (passenger) strand being blocked from RISC uptake to favor antisense (guide) strand loading and reduce passenger strand-induced off-targets. However, the majority of siRNA off-targets are driven by the seed region of the guide strand. ON-TARGETplus is modified within its seed region to destabilize miRNA-like activity and improve specificity to the desired target for potent knockdown.
siRNA designs with low-frequency seed regions ensure fewer off-targets
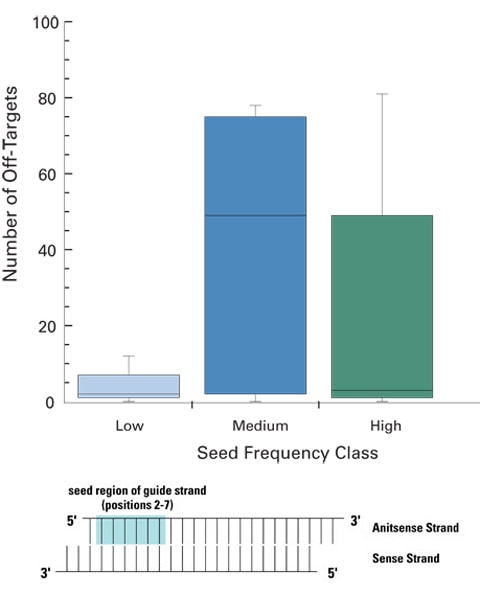
ON-TARGETplus siRNA designs leverage sophisticated bioinformatics to reduce the likelihood of miRNA-like off-targets from high-frequency or highly conserved miRNA seed regions. siRNAs with low seed frequency have a significantly lower number of off-targets than siRNAs with medium or high frequency seeds. Five siRNAs with low, medium, or high frequency seed regions were transfected into HeLa cells and their associated off-target signatures assessed via global expression profiling (Agilent 22K platform). siRNA sequences were constant at positions 1 and 8-19, only the seed regions (positions 2-7) were altered.
Low frequency seeds: < 350 occurrences in the HeLa transcriptome
Medium frequency: 2500-2800 occurrences
High frequency: >3800 occurrences
ON-TARGETplus 2.0 maintains high performance of target gene knockdown set by ON-TARGETplus

U2OS or HCT-116 cells were transfected with 25 nM ON-TARGETplus or ON-TARGETplus 2.0 siRNAs targeting the AHCY (left) or PRDX1 (right) genes, or a non-targeting control (NTC) siRNA with 0.2 µL/well DharmaFECT 4 (U2OS) or DharmaFECT 2 (HCT-116). After 48 hours, total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene knockdown was observed with both ON-TARGETplus and ON-TARGETplus 2.0 siRNAs. n = 2 biologically independent samples per group.
ON-TARGETplus and ON-TARGETplus 2.0 siRNAs potently deplete TP53 mRNA expression with minimal impacts on global gene expression

U2OS cells were transfected with 25 nM ON-TARGETplus or ON-TARGETplus 2.0 siRNAs targeting the TP53 gene, or a non-targeting control (NTC) siRNA with 0.2 µL/well DharmaFECT 4. A. After 48 hours, total RNA was isolated and prepped for RNAseq or RT-qPCR. A. Data plotted as a histogram showing the distribution of global transcriptional expression, represented as transcripts per million (TPM). Strong downregulation of TP53 mRNA expression was observed in all four siRNA treatment groups, denoted on the x-axis and represented as log2 transformed TPM fold change while global gene expression was largely unchanged relative to the corresponding NTC siRNA. n = 2 biologically independent samples per group. B. Relative gene expression was measured using RT-qPCR. The relative expression of TP53 was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene knockdown was observed with both ON-TARGETplus and ON-TARGETplus 2.0 siRNAs. n = 2 biologically independent samples per group.
ON-TARGETplus 2.0 mitigates newly discovered off-target effects arising from transcriptome expansion

U2OS or HEK293T cells were transfected with 25 nM ON-TARGETplus or ON-TARGETplus 2.0 siRNAs targeting the YY1AP1 gene, or a non-targeting control (NTC) siRNA with 0.2 µL/well DharmaFECT 1 (HEK293T) or DharmaFECT 4 (U2OS). After 48 hours, total RNA was isolated and prepped for RNAseq or RT-qPCR. A. Data plotted as a histogram showing the distribution of global transcriptional expression in U2OS cells, represented as transcripts per million (TPM). Strong downregulation of YY1AP1 mRNA expression was observed in all four siRNA treatment groups, denoted on the x-axis and represented as log2 transformed TPM fold change while global gene expression was largely unchanged relative to the corresponding NTC siRNA. Notably, ON-TARGETplus siRNAs also depleted expression of the off-target gene GON4L whereas ON-TARGETplus 2.0 siRNAs maintained high specificity and did not affect GON4L expression. n = 2 biologically independent samples per group. B. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene knockdown was observed with both ON-TARGETplus and ON-TARGETplus 2.0 siRNAs, however the ON-TARGETplus siRNA was found to also reduce expression of an off-target gene, GON4L. The ON-TARGETplus 2.0 siRNAs maintained high specificity and did not affect GON4L expression. n = 2 biologically independent samples per group.
- A. L. Jackson et al., Position-specific chemical modification increases specificity of siRNA-mediated gene silencing. RNA. 12(7), 1197-1205 (July 2006).
- A. Birmingham et al., 3'-UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods. 3(3), 199-204 (March 2006).
- E. M. Anderson et al., Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 14(5), 853-861 (May 2008).
Please see our full FAQ page for all commonly asked questions about our products.
- What is the difference between ON-TARGETplus legacy and OTP 2.0?
- How does OTP 2.0 reduce off-target effects?
- Are OTP 2.0 siRNAs guaranteed to knock down my target gene?
- In what formats are OTP 2.0 siRNAs available?
- Can OTP 2.0 be used in high-throughput or automated platforms?
- Are OTP 2.0 siRNAs available for multiple species?
- How does OTP 2.0 support genome-wide and gene-family studies?
- Can OTP 2.0 be used alongside Edit-R CRISPR reagents?
OTP 2.0 is a next-generation upgrade of the original ON-TARGETplus siRNA. It features improved design algorithms, continuous transcriptome realignment, and updated sequence ranking to enhance specificity, minimize off-target effects, and ensure consistent gene knockdown. In contrast, legacy OTP designs were based on earlier transcriptome data and do not include these ongoing optimizations.
Each siRNA is designed using updated transcriptome data and advanced ranking algorithms that prioritize specificity. Seed-region optimization and refined design parameters help minimize unintended interactions.
Yes. OTP 2.0 siRNAs are backed by a guaranteed gene knockdown standard, ensuring consistent and dependable performance when used under recommended conditions.
OTP 2.0 is offered as individual siRNAs or as SMARTpool reagents (a blend of four different siRNAs designs targeting the same gene). Plate-based formats are available to support screening workflows.
Absolutely. OTP 2.0 siRNAs are available in formats compatible with automated systems, including plates suitable for high-throughput applications.
Yes. Pre-designed OTP 2.0 siRNAs are available for human, mouse, and rat gene targets.
OTP 2.0 collections cover whole-genome libraries as well as focused sets for commonly studied gene families, enabling both broad and targeted screening approaches. In addition, cherry-pick libraries can be built for desirable targets.
Yes. OTP 2.0 serves as an ideal orthogonal method to validate CRISPR screening hits or complement gene knockout studies with gene knockdown testing.
Application notes
Product inserts
Protocols
Safety data sheets
Selection guides
Related Products
Negative control pool of four siRNAs designed and microarray tested for minimal targeting of human, mouse or rat genes. ON-TARGETplus modifications reduce potential off-targets. Recommended for determination of baseline cellular responses in RNAi experiments.
Validated positive control pool of four siRNAs targeting GAPD gene in human, mouse, or rat. Includes patented ON-TARGETplus modifications to minimize off-target effects. Useful for determination of optimal RNAi conditions.
Concentrated buffer solution recommended for resuspension and long-term storage of any short, double-strand, or single-strand synthetic RNA molecule. Dilute with RNase-free water prior to use.
Catalog ID:B-002000-UB-100
Unit Size:100 mL
$99.00
An aliquot of each of the four DharmaFECT formulations for siRNA/microRNA transfection optimization studies
Catalog ID:T-2005-01
Unit Size:0.2 mL
