While genome engineering is widely ascribed with the capacity to knock out genes of interest, new adaptations of these same molecular building-blocks are reshaping the way researchers think about the opposite: overexpression experiments. This technique allows recapitulation of the native gene expression patterns in a manner previously unobtainable using traditional overexpression-plasmid methods. For CRISPR activation (CRISPRa), the guide RNA forms a complex with a nuclease-deactivated Cas9 (dCas9), which is in turn fused to three transcriptional activators (comprising the VPR system). The machinery then acts upstream of the transcription start site to up-regulate expression of a target gene (Figure 1). CRISPRa provides a new set of tools to identify gene function that might otherwise go undetected using loss-of-function studies through gene knockdown or knockout.
The Dharmacon Edit-R CRISPRa system
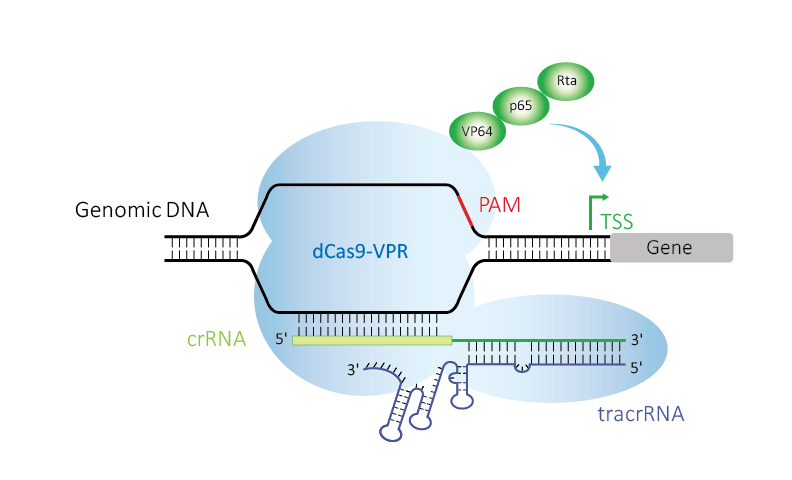 Figure 1. Edit-R CRISPRa uses deactivated Cas9 (dCas9) fused to the transcriptional activators VP64, p65, and Rta (VPR), guided by crRNA:tracrRNA upstream of a transcriptional start site (TSS) to up-regulate expression of a target gene.
Figure 1. Edit-R CRISPRa uses deactivated Cas9 (dCas9) fused to the transcriptional activators VP64, p65, and Rta (VPR), guided by crRNA:tracrRNA upstream of a transcriptional start site (TSS) to up-regulate expression of a target gene.
Previously, we have demonstrated that synthetic crRNAs can be successfully used to perform experiments in cells stably expressing dCas9-VPR. However, the constitutive expression or extended timeline needed to create a stable cell line may not be desired for every application. Here we examined the ability to use the synthetic crRNA approach with dCas9-VPR mRNA - alleviating the need to use lentivirus altogether.
We demonstrate that:
- Transient delivery of dCas9-VPR mRNA and synthetic guide in multiple cell lines results in robust activation – CRISPRa is possible in any cell line that is amenable to lipid-mediated or electroporation delivery methods.
- Using EGFP-tagged or puromycin-tagged dCas9-VPR mRNA enables antibiotic or fluorescent selection – CRISPRa can be improved by enrichment methods when delivery is less than optimal.
Transcriptional activation by transient transfection of dCas9-VPR mRNA and synthetic guide RNA
To demonstrate that we can get transcriptional activation without the need to generate a stable-expressing dCas9-VPR cell line, we used DharmaFECT Duo to co-transfect Edit-R dCas9-VPR mRNA, Edit-R synthetic tracrRNA, and Edit-R CRISPRa crRNA pools targeting the promotor region of either POU5F1 or IL1R2. We observed robust activation of approximately 1,000-fold (Figure 2).
CRISPRa gene activation by co-transfection of dCas9-VPR mRNA and synthetic guide RNA
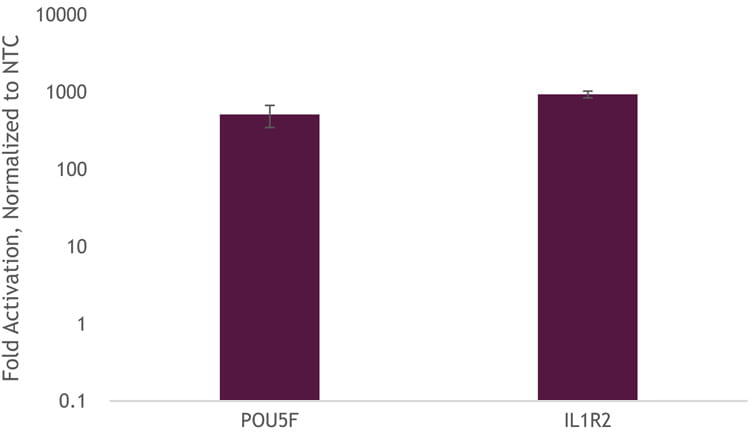 Figure 2. U2OS cells were plated at 10,000 cells per well, in clear 96-well plates. After 24 hours, cells were co-transfected with dCas9-VPR mRNA (200 ng, Cat #CAS12024), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting either POU5F1 (25 nM Cat #P-019591-01-0005) or IL1R2 (25 nM, Cat #P-007960-01-0005), or non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (0.25 µg/well, Cat #T-2010-01). Cells were harvested at 48 hours post-transfection and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
Figure 2. U2OS cells were plated at 10,000 cells per well, in clear 96-well plates. After 24 hours, cells were co-transfected with dCas9-VPR mRNA (200 ng, Cat #CAS12024), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting either POU5F1 (25 nM Cat #P-019591-01-0005) or IL1R2 (25 nM, Cat #P-007960-01-0005), or non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (0.25 µg/well, Cat #T-2010-01). Cells were harvested at 48 hours post-transfection and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
To test whether this same activation could be demonstrated in cells where electroporation is necessary for delivery, we co-electroporated K-562 and THP-1 cells with dCas9-VPR mRNA, synthetic tracrRNA and crRNA pools targeting the promotor region of TTN and see strong activation (Figure 3). We also observe robust activation of TTN for K-562 and THP-1 cells co-electroporated with EGFP or puro tagged mRNA, which enables enrichment and a wider array of applications.
CRISPRa gene activation by electroporation of dCas9-VPR mRNA and synthetic guide RNA
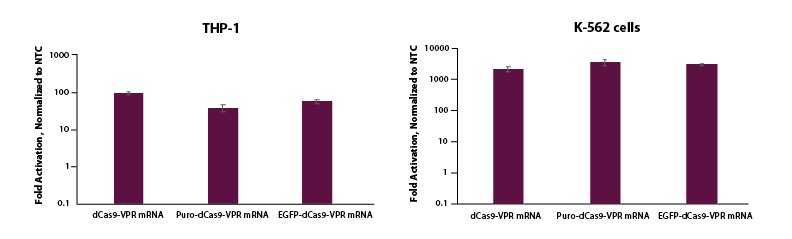 Figure 3. THP-1 and K-562 cells were electroporated using the Lonza 2b system with either dCas9-VPR mRNA (5 µg, Cat #CAS12024), Puro dCas9-VPR mRNA (5 ug, Cat #CAS12026) or EGFP dCas9-VPR mRNA (5 ug, Cat #CAS12025), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting TTN (5 uM, Cat #P-005395-01-0005) or Non-targeting control (NTC, 25 nM, Cat #U-009500-10-05). Cells were harvested at 48 hours post-transfection and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
Figure 3. THP-1 and K-562 cells were electroporated using the Lonza 2b system with either dCas9-VPR mRNA (5 µg, Cat #CAS12024), Puro dCas9-VPR mRNA (5 ug, Cat #CAS12026) or EGFP dCas9-VPR mRNA (5 ug, Cat #CAS12025), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting TTN (5 uM, Cat #P-005395-01-0005) or Non-targeting control (NTC, 25 nM, Cat #U-009500-10-05). Cells were harvested at 48 hours post-transfection and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
dCas9-VPR mRNA allows for enrichment using antibiotics or fluorescence
EGFP- or puro-tagged dCas9-VPR mRNA can be used to select for enrichment in gene activation experiments where the cell line may have low delivery efficiency, or in cases where optimization of delivery conditions is not possible.
For EGFP dCas9-VPR mRNA, we co-transfected EGFP dCas9-VPR mRNA, synthetic tracrRNA, and crRNA pools targeting the promotor region of IL1R2. Following FACS sorting we observed significant enrichment for the top 10% sorted cells when compared to dim and unsorted cell populations (Figure 4).
EGFP fluorescence can be used to select for cells with EGFP dCas9-VPR mRNA and enrich for gene activation
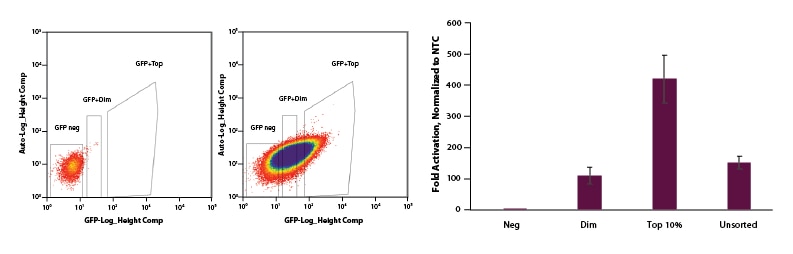
Figure 4. U2OS cells were plated at 200,000 cells per well in clear 6-well plates. After 24 hours, cells were co-transfected with EGFP dCas9-VPR mRNA (2 µg, Cat #CAS12025), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting IL1R2 (25 nM, Cat #P-007960-01-0005) or Non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (2 µg/well, Cat #T-2010-01). A: At 24 hours post-transfection, cells were trypsinized and FACS was performed and cells were sorted into three categories: Negative, Dim, and Top 10%. Cells were replated in 6-well dishes and allowed to recover. B: After 24 hours, total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control. The Negative population of sorted cells shows no activation. The Dim and Unsorted populations both showed ~120-fold activation and our Top 10% population resulted in 450-fold activation.
We further examined if selection and enrichment could be achieved using our Edit-R Puro dCas9-VPR mRNA. Similarly, we co-transfected cells with Edit-R Puro dCas9-VPR mRNA, Edit-R synthetic tracrRNA, and Edit-R crRNA pools targeting the promotor region of either IL1R2, TFAP2C, or TTN. Following a 24-hour selection with puromycin supplemented media, we observed significant enrichment or gene activation in our puromycin selected population (Figure 5).
Puromycin can be used to select for cells with Puro dCas9-VPR mRNA and enrich for gene activation
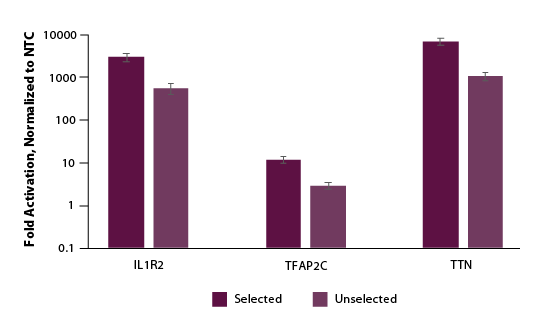 Figure 5. U2OS cells were plated at 200,000 cells per well in clear 6-well plates. After 24 hours, cells were co-transfected with Puro dCas9-VPR mRNA (2 µg, Cat #CAS12026), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting either POU5F1 (25 nM Cat #P-019591-01-0005), IL1R2 (25 nM, Cat #P-007960-01-0005), TFAP2C (25 nM, Cat #P-005238-01-0005), TTN (25 nM, Cat #P-005395-01-0005) or non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (2 µg/well, Cat #T-2010-01). At 24 hours post-transfection, 2 µg/ml of puromycin growth media was added to the cells and a duplicate plate received normal growth media. At 48 hours post-transfection a crystal violet assay was performed and images captured to assess viability and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control. For all genes tested, we observed 3- to 5-fold enrichment of activation when compared to unselected samples.
Figure 5. U2OS cells were plated at 200,000 cells per well in clear 6-well plates. After 24 hours, cells were co-transfected with Puro dCas9-VPR mRNA (2 µg, Cat #CAS12026), synthetic tracrRNA (25 nM, Cat #U-002005-05), and pooled CRISPRa crRNA targeting either POU5F1 (25 nM Cat #P-019591-01-0005), IL1R2 (25 nM, Cat #P-007960-01-0005), TFAP2C (25 nM, Cat #P-005238-01-0005), TTN (25 nM, Cat #P-005395-01-0005) or non-targeting control (NTC, 25 nM, Cat #U-009500-10-05) using DharmaFECT Duo (2 µg/well, Cat #T-2010-01). At 24 hours post-transfection, 2 µg/ml of puromycin growth media was added to the cells and a duplicate plate received normal growth media. At 48 hours post-transfection a crystal violet assay was performed and images captured to assess viability and total RNA was isolated. Relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control. For all genes tested, we observed 3- to 5-fold enrichment of activation when compared to unselected samples.
Conclusions
dCas9-VPR mRNA can be used in both lipid-mediated transfection and electroporation delivery to achieve transcriptional activation in a CRISPRa system. Furthermore, EGFP dCas9-VPR mRNA and Puromycin dCas9-VPR mRNA can be used to select and enrich for gene activation.
These methods presented are broadly applicable as a strategy to up-regulate the expression for any gene in difficult-to-transfect cell lines, or in laboratories where generating stably expressing dCas9-VPR cell lines may be technically difficult or undesirable.
Finally, these advances in technology open the door for systems biology experiments previously unfeasible, such as large-scale overexpression screening or overexpressing multiple genes in the same cell.
Additional Resources
Edit-R CRISPR Activation Reagents
- Synthetic & lentiviral reagents for endogenous gene overexpression
Edit-R CRISPRa Transcriptional Activation System
- Learn more about our Edit-R CRISPRa reagents
