A method for uncovering off-target effects
RNAi screens with arrayed siRNA collections are a powerful and popular discovery tool, but are plagued by the endogenous microRNA-like silencing of unintended targets due to activity of the siRNA seed region (nt 2-8 of the antisense strand). In fact, there has been demonstration that introduction of siRNAs without a target sequence can reproduce phenotypes simply through activity of their seed region, and that point mutations within the seed abolished this activity1.
While there are many powerful analytical approaches to determine if a phenotype is real or a seed-based off-target2, 3, there is also a bench-level approach to control for this activity: Seed sibling controls.
What is a seed sibling?
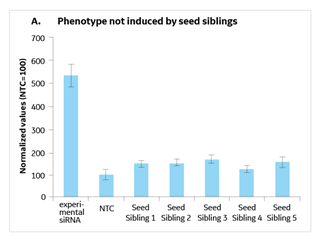
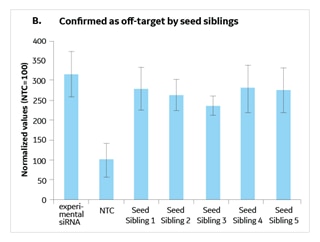
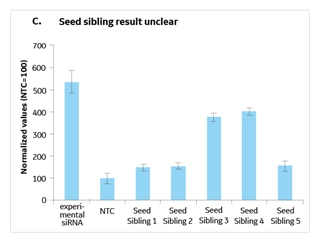
A seed sibling is a siRNA that has no perfect match to transcripts in a species of interest, but whose seed is a perfect match to an experimental siRNA that gives a phenotype (Figure 1). By running a validation study with experimental siRNAs side-by-side with 4-5 of its seed siblings, you can determine if the seed alone is active in initiating a phenotype (since it has no known targets) thereby indicating that the phenotype is likely due to a seed-mediated off-target effect (Figure 2). Alternatively, if the seed siblings do not generate a phenotype, there is good indication that the hit was due to knockdown of the intended target and not a seed-based off-target effect.
| Experimental siRNA (targets a human transcript): | Seed sibling control (no human target): | |
|---|---|---|
| Sense: Antisense: | 5’-CAACAGCCUUCAAGCCAGU-oo oo-GUUGUCGGAAGUUCGGUCA-5’ | 5’-CAACAGUAGUCAAGCCAGA-oo oo-GUUGUCAUCAGUUCGGUCU-5’ |
 |
 |
 |
Ordering information
Obtaining seed sibling control siRNAs can be accomplished with a lookup tool as part of the Dharmacon Cherry-pick Plater. When a researcher provides a list of experimental siRNAs to validate (either a Dharmacon catalog number or the sense/target sequence), the tool will return a list of catalog siRNAs that have the same seed region as the indicated experimental siRNA, but with no perfect match target in the species of interest. See Figure 3 for an example. Since this approach uses existing Dharmacon catalog products, these reagents are immediately available as there is no custom synthesis required.
The selected seed sibling siRNAs can then be automatically loaded into the Cherry-pick Plater to generate an arrayed plate with the desired quantity per well, catalog controls, and plate layout.
| Experimental siRNA (Dharmacon catalog # or sense sequence, provided by researcher) | Seed siblings (returned by the online tool) |
|---|---|
| D-026306-02 (targets human LZTS2) | D-051682-04 (targets mouse Angel1) D-117390-01 (targets rat LOC100912087) D-061580-04 (targets mouse Dync1li2) D-058071-17 (targets mouse Ssxa1) |
References:
- A. Franceschini, R. Meier, et al. Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens PNAS 111(12); 4548-4553, 2014
- E. Buehler et. al., Common Seed Analysis to Identify Off-Target Effects in siRNA Screens J. Biomol Screening 17 (3) 370-378, 2012
- F. Sigoillot, S. Lyman et. al., A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nature Methods 9, 363–366 (2012)
Order Products
Ready to get some seed sibling controls?
Go to the Cherry-pick plater and get started!
RNAi Screening Libraries
Small gene families to genome-wide collections provide flexible, convenient and high-confidence screening results
Helpful Resources
Strategies for Improving RNAi Screening Success: Using a Ubiquitin-EGFP Assay to Identify Druggable Genes Required for Proteasome Function
Data from an example RNAi screen is presented, along with recommendations for best practices
