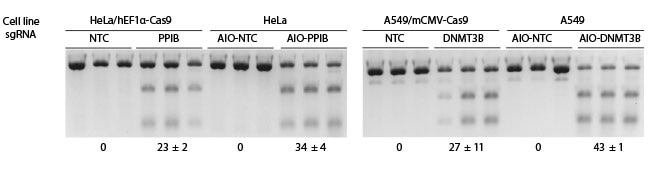
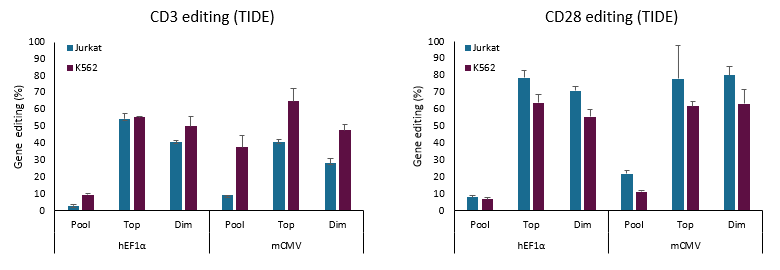
CRISPR workflows are simplified with Dharmacon All-in-one reagents. The All-in-one platform combines the CRISPR nuclease and single guide RNA expression into a single lentiviral packaged vector – for the simplest gene knockout/modulation workflow available. One reagent CRISPR for any human or mouse gene.
| All-in-one benefits | Ideal for |
|---|---|
|
|
|
|
|
|
|
|
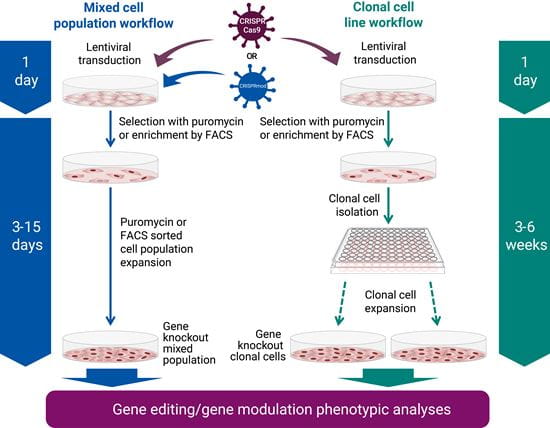
All-in-one CRISPR gene knockout, activation, and interference workflow
 The power of the All-in-one system lies in the simplicity of it's workflow
The power of the All-in-one system lies in the simplicity of it's workflow
To transduce, simply thaw the reagent and add the appropriate amount to your cells. Then easily select for targeted cells using puromycin or enrich using FACS.
When using puromycin selection, growth medium is replaced with puromycin selection medium 24-48 hours post-transduction. Cells are then monitored daily until growing normally in the selection medium. Once growth is stable, isolation and/or expansion of the targeted cell population(s) may occur.
Alternatively, using the EGFP fluorescent reporter option offers the ability to enrich for edited cells via FACS selection in as few as 72 hours post-transduction. The FACS enrichment workflow is especially useful for short-lived cell types, such as primary cells.
For CRISPRa and CRISPRi, we recommend the mixed cell population workflow.
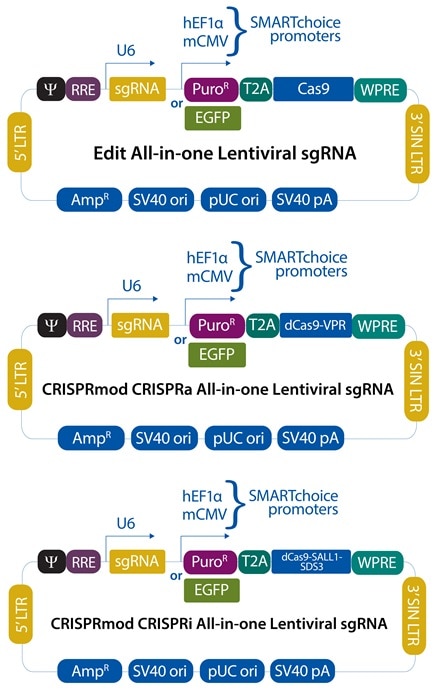
CRISPR knockout all-in-one lentiviral system
CRISPRko single reagent
CRISPR sgRNA + Cas9 in one vector for easy gene editing
CRISPRko whole genome library
All-in-one high titer pooled screening library covering the entire human genome
CRISPR activation all-in-one lentiviral system
CRISPRa single reagent
CRISPRa sgRNA + dCas9-VPR in one vector for easy gene activation
CRISPRa whole genome library
CRISPRa all-in-one high titer pooled screening library covering the entire human genome
CRISPR interference all-in-one lentiviral system
CRISPRi single reagent
CRISPRi sgRNA + dCas9- SALL1-SDS3 in one vector for easy transcriptional repression
CRISPRi whole genome library
CRISPRi All-in-one high titer pooled screening libraries covering the entire human genome
The DharmaconTM All-in-one systemThe All-in-one system offers the simplest CRISPR workflow available – a single reagent, easy delivery and straightforward selection. Single reagentUsing a single vector to express both CRISPR nuclease and gene-specific guide RNA eliminates the need to perform sequential transduction and selection steps. Easy deliveryLentiviral packaging allows for easy transduction into nearly any cell type. Eliminating the need for toxic transfection reagents or electroporation techniques makes the All-in-one system particularly useful for genome editing and transcriptional modulation in difficult-to-transfect or primary cell types. Transduction-ready reagents also eliminate any cloning or in vitro transcription steps. Straightforward selectionThe choice of a puromycin resistance gene or EGFP marker allows for straightforward selection of cells that have successfully integrated the vector. EGFP is recommended for rapid enrichment of targeted cells, as FACS analysis may be performed as soon as fluorescence is expressed.
Promoter optionsChoice of promoters allows you to select the most active promoter in your cell line. |
 |
Efficient transcriptional activation with CRISPRmod CRISPRa All-in-one Puro-dCas9-VPR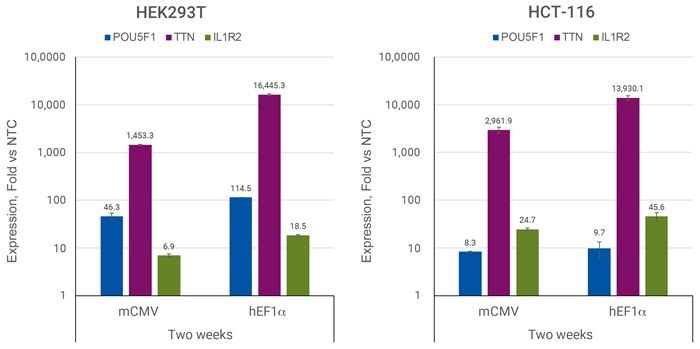
HEK293T or HCT-116 cells were plated in 96-well plates at 20,000 or 5,000 cells/well, respectively, and transduced with 0.3 MOI CRISPRa all-in-one virus containing sgRNAs targeting POU5F1, TTN, IL1R2, or Non-targeting control #2 (NTC2). Cells were selected with 2.5 µg/µL puromycin for 5 days, allowed to expand, and harvested two weeks post-transduction. Relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control.
Increasing multiplicity of infection (MOI) improves target activation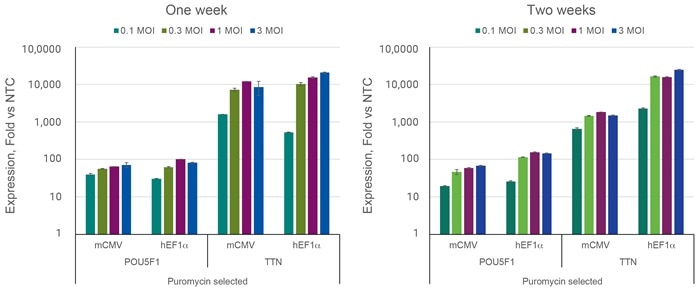
HEK293T cells were plated in 96-well plates at 20,000 cells/well and transduced with denoted MOIs of CRISPRa all-in-one virus containing sgRNAs targeting POU5F1, TTN, or NTC2. Cells were selected with 2.5 µg/µL puromycin for 5 days, allowed to expand, and harvested one or two weeks post-transduction. Relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control.
Induction of OCT4 protein expression in unsorted U2OS cells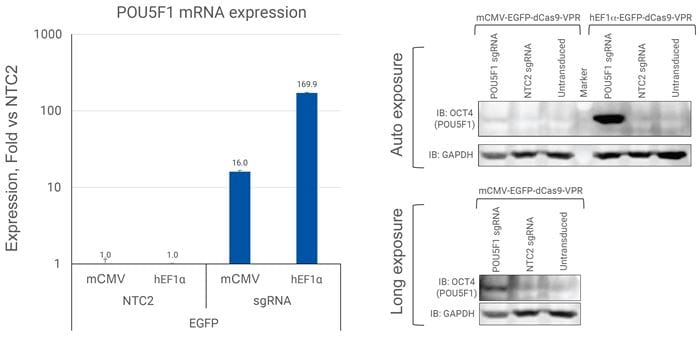
U2OS cells were plated in 96-well plates at 10,000 cells/well and transduced with 0.5 MOI EGFP CRISPRa All-in-one virus containing sgRNAs targeting POU5F1 or NTC2. Cells were passaged and expanded into 96-well and 6-well plates. RNA was extracted from cells cultured in 96-well plates one-week post-transduction. Relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control.
Activation of OCT4 protein in unsorted U2OS cells by EGFP mCMV CRISPRa All-in-one viral particles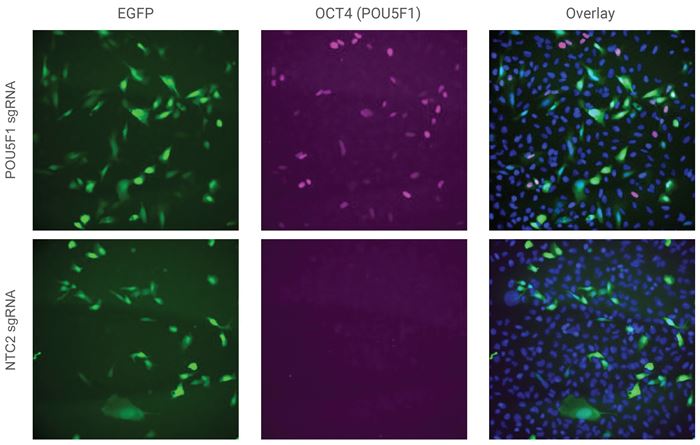
U2OS cells were plated in 96-well plates at 10,000 cells/well and transduced with 0.5 MOI EGFP CRISPRa All-in-one virus containing sgRNAs targeting POU5F1 or NTC2.
|
Overexpress your target gene at the transcriptional levelFind All-in-one lentiviral sgRNA for CRISPR activation for any human gene
|
Efficient transcriptional repression with CRISPRmod CRISPRi All-in-one system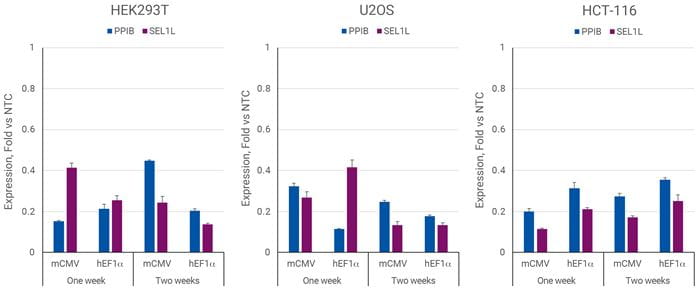
HEK293T, U2OS, or HCT-116 cells were plated in 96-well plates at 20,000, 10,000, or 5,000 cells/well and transduced with 0.3 MOI CRISPRi all-in-one virus containing sgRNAs targeting PPIB, SEL1L, or NTC2. Cells were selected with 2.5 µg/µL puromycin for 5 days, allowed to expand, and harvested at denoted time points post-transduction. Relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control.
With puromycin selection transcriptional repression is improved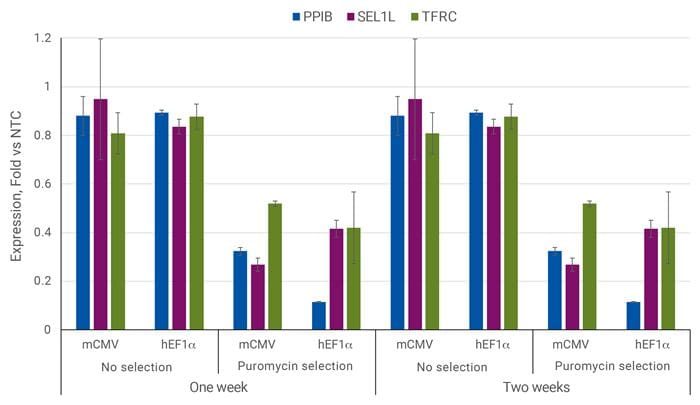
U2OS cells were plated in 96-well plates at 10,000 cells/well and transduced with 0.3 MOI CRISPRi all-in-one virus containing sgRNAs targeting PPIB, SEL1L, TFRC, or NTC2. Cells were selected with 2.5 µg/µL puromycin (where applicable) for 5 days, allowed to expand, and harvested one week or two weeks post-transduction. Relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control.
Increased transcriptional repression in EGFP positive Jurkat cells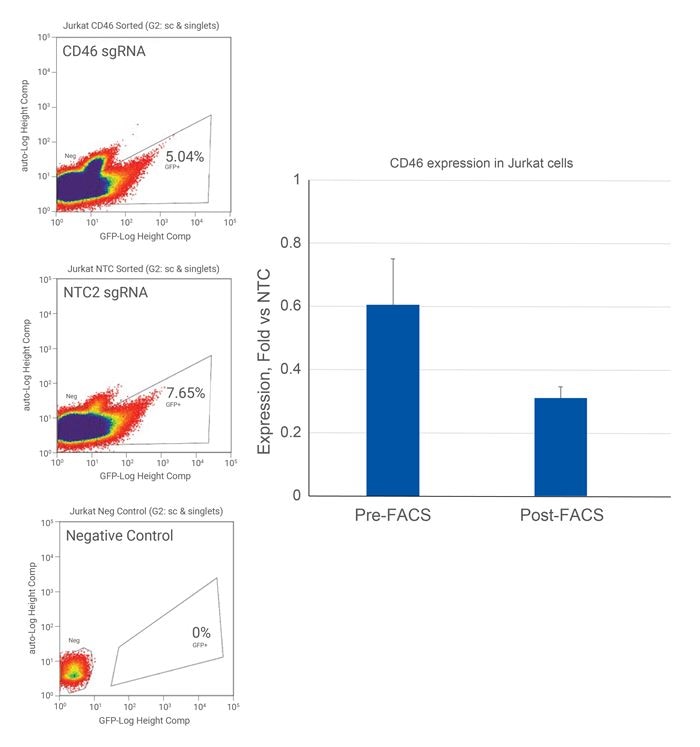
Jurkat cells were plated at 25,000 cells/well and transduced with 0.3 MOI CRISPRi all-in-one virus containing sgRNAs targeting CD46 or NTC2 (CRISPRi). Cells were cultured for one week and harvested for mRNA extraction (1-week post-transduction, Pre-FACS) or cultured for 3 weeks, washed in PBS, and resuspended in FACS buffer (1X PBS containing 10 mM Tris-HCl - pH 7.5, 25 mM HEPES, and 1% FBS). EGFP positive cells were sorted via FACS into 24 well plates containing ~50,000 EGFP positive cells per well. Cells were expanded for 1 week, and RNA was harvested at four weeks post-transduction (Post-FACS). Relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control. |
Knock down your target gene at the transcriptional levelFind All-in-one lentiviral sgRNA for CRISPR interference for any human gene
|
Pooled CRISPR screening libraries are essential for genome-level cell system interrogation, addressing numerous biological questions such as drug and pathogen mechanisms and gene essentiality. However, delivering lentiviral CRISPR components sequentially is challenging, especially in difficult-to-transfect or primary cell types. Additionally, packaging vectors that express both Cas9 effectors and gene-specific single guide RNA (sgRNA) into high-titer lentiviral libraries is taxing, especially with large deactivated Cas9 (dCas9) fusions needed for CRISPR activation (CRISPRa) or CRISPR interference (CRISPRi).
Our high-titer, uniformly distributed Dharmacon™ All-in-one lentiviral pooled CRISPR libraries overcome these challenges.
High quality pooled screening begins with rigorous lentiviral pooled library production
All three varieties of Dharmacon all-in-one lentiviral pooled libraries are shown having high recovery of input sgRNAs (98%+) and even distributions as verified by next-generation sequencing (NGS).

Robust functional titers with Dharmacon™ all-in-one lentiviral pooled libraries
Titer verification of an all-in-one pooled library and library-matched delivery controls with two methods. All our pooled libraries are functionally tittered. Comparing the methods one can observe that p24 tittering overestimates the actual value.