- Products
- Engineered cell lines
- HAP1 knockout cell lines
HAP1 knockout cell lines
The single largest bank of isogenic cell lines with over 7,500 cell lines to choose from and trusted by academia, biotech, and pharma research labs. Cell models to suit any application, from TIDVAL to antibody validation, delivered in as few as 10 days.
HAP1 knockout cell lines
1Start Here
2Choose
Advantages of HAP1 knockout cell lines
- Easily characterize knockout of your gene-of-interest in a biological context
- Ready-made knockout cell lines are available for all non-essential human genes
- Editing has been confirmed by Sanger sequencing of genomic DNA
- Two independent knockout products for each gene, allowing second-clone analysis if more than one data point is required
Proven models
Our catalog of cell lines has become a gold-standard resource in antibody validation, cancer research, and early drug-discovery research. Horizon was recently recognized by CiteAb as the Cell Line Supplier to Watch in Cancer Research for 2024. Read more about the CiteAb award.
in antibody validation, cancer research, and early drug-discovery research. Horizon was recently recognized by CiteAb as the Cell Line Supplier to Watch in Cancer Research for 2024. Read more about the CiteAb award.
Why are HAP1 cells an ideal option for gene-edited cell line models?
Our HAP1 cell line database represents the synergy of two great systems - the amenability of human haploid HAP1s to gene editing and CRISPR-Cas9 technology. HAP1 is a human near-haploid cell line derived from the chronic myelogenous leukemia (CML) cell line KBM-7. KBM-7 cells were derived from a male patient and lack a Y chromosome. Therefore, haploid HAP1 cells contain a single X chromosome. For more details on how the HAP1 cell line was created and their full genotype, please see the references tab.
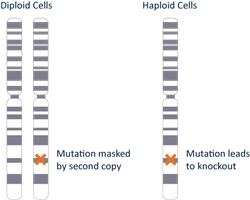
HAP1 cell line properties include:
- Sustained growth at single cell dilutions
- Ease of transfection
- Rapid doubling time
- Not selected for ploidy status
- Will have haploid and diploid populations
The majority of commonly used cell lines are diploid or even have more than two copies of any particular allele. When gene editing for functional reduction mutations, each allele has to be modified for the phenotype to be expressed. The HAP1 cell line has the advantage of having one copy of each gene meaning you can be sure the edited allele will not be masked by addition alleles. With passaging, HAP1 cells will spontaneously diploidize over time. However, the knockout will not be affected by diploidization as it is a duplication of the edited allele that gives rise to the diploid state.
See our HAP1 publication list to see how the cell line has been applied for a wide range of biological processes and assays, such as DNA damage repair pathway, stress responses, disease modeling as well as antibody validation.
Limited use agreement
All cell lines engineered by Horizon Discovery are purchased under the terms of our Limited Use Label License.
We are confident that HAP1 and Cancer-related Ready-to-go cell lines are the ideal product to further your research
However, if you are not satisfied with the performance of any of our HAP1 and Cancer-related Ready-to-go cell lines, please contact us at technical@horizondiscovery.com and we will work diligently with you to resolve any problems, as detailed in our terms and conditions of sales.
References
For more information about how the HAP1 cell line was generated, see:
- Kotecki, M., Reddy, P. S. & Cochran, B. Isolation and Characterization of a Near-Haploid Human Cell Line. Experimental Cell Research 252, 273–280 (1999). DOI: 10.1006/excr.1999.4656
- Carette, J. E. et al. Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens. Science 326, 1231–1235 (2009). DOI: 10.1126/science.1178955
- Carette, J. E. et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature 477, 340–343 (2011). DOI: 10.1038/nature10348
- Essletzbichler, P. et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res 24, 2059–2065 (2014). DOI: 10.1101/gr.177220.114
- Dong, M. et al. DAG1 mutations associated with asymptomatic hyperCKemia and hypoglycosylation of α-dystroglycan. Neurology 84, 273–279 (2015). DOI: 10.1212/WNL.0000000000001162
- Kravtsova-Ivantsiv, Y. et al. KPC1-mediated ubiquitination and proteasomal processing of NF-κB1 p105 to p50 restricts tumor growth. Cell 161, 333–347 (2015). DOI: 10.1016/j.cell.2015.03.001
- Lackner, D. H. et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun 6, 10237 (2015). DOI: 10.1038/ncomms10237
