Most research projects for functional analysis are centered on protein-coding genes. After all, it’s proteins (< 2% of the genome)1 that send and receive signals, serve as structural building blocks, and regulate nearly every key biological function. But what about the 70-90% of the human genome that is transcribed, but not translated into a protein 2,3. Past and ongoing studies have demonstrated the immensely important role of different types of regulatory noncoding RNAs (ncRNAs) in development and disease4.
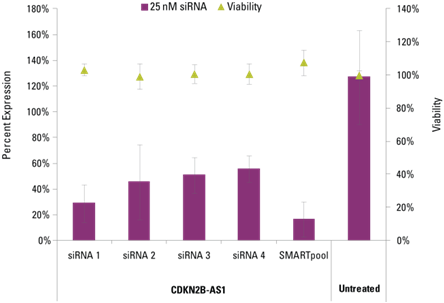
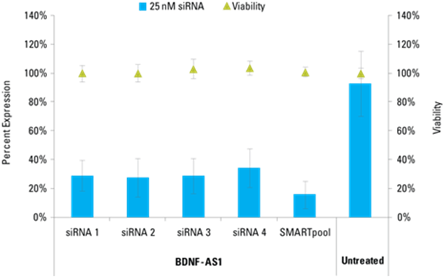
Lincode siRNAs effectively knockdown target lncRNAs
Detection of effective lncRNA knockdown following application of Lincode siRNA reagents
microRNA
microRNAs are short, noncoding RNAs that modulate gene expression in a post-transcriptional manner by binding to the 3’-UTR of a targeted transcript by imperfect base pairing of the microRNA seed region (bases 2-7 or 2-8). This mechanism uses endogenous RNAi machinery to cleave and degrade the target transcript or inhibit its translation, thereby carrying out its regulatory function. The identification of microRNA’s role in development, cancer, cardiovascular, and other diseases5 as well as their potential as therapeutics continue to support the functional study of these small regulatory molecules6.
Long noncoding RNA
Long noncoding RNAs (lncRNAs) have garnered attention as another species of regulatory RNA with key roles in development, epigenetics, regulation of transcription, and other essential biological processes in addition to being linked to diseases like Alzheimer’s disease7, breast cancer8, and melanoma9. Identification and functional study of lncRNAs are essential to interpretation of genome wide association studies, and play a valuable role as highly sensitive biomarkers as well as potential therapeutic targets. Uncovering their biological roles is fundamental to understanding the molecular basis of disease4.
Challenges in studying ncRNAs
Although the tools and techniques for the study of protein-coding genes are very well characterized, different analytical approaches are required for the study of ncRNAs because of their distinct mechanisms and mode of action.
Expression
The overall expression levels of ncRNAs vary widely. While microRNAs show a range of expression, many microRNA and lncRNAs are expressed at much lower levels than protein-coding genes, and their temporal expression in highly specific tissues at certain points in time raises challenges to research whose goal is to uncover their specific function.
Detection
Since microRNAs and lncRNAs can have low expression levels, the isolation and quantification of small RNA species is critical and detection technologies often require amplification. As such, the result of lncRNA knockdown or a microRNA inhibition experiment might be challenging to interpret without a characterized target or known phenotype.
Target identification
The mechanism of microRNAs is well understood and in silico analysis and experimental approaches allow for identification of their targets. Since the role of a lncRNA is to act upon other cellular components including RNAs, DNA or protein4, it is essential to understand the target of a ncRNA to uncover its function. New next-generation sequencing, biochemical and in silico techniques are being utilized to address mechanistic questions in this challenging field.
Tools for modulation of regulatory RNA
In support of the interest in regulatory ncRNA, we have assembled a toolkit for effective modulation of genes encoding characterized lncRNAs and microRNAs. The Dharmacon product range includes synthetic siRNA to target lncRNAs, synthetic microRNA tools for suppression or up-regulation of endogenous mature microRNA function, and constitutive or inducible microRNA expression vectors. In addition, crRNA or sgRNA can be utilized to knockout or modify ncRNAs. Together, these tools can be used to uncover the roles of regulatory RNAs in biological pathways and networks.
References
- J.E. Wilusz, H. Sunwoo, D.L. Spector, Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23, 1494-1504 (2009).
- J. T. Kung, D. Colognori, J. T. Lee, Long noncoding RNAs: past, present, and future. Genetics. 193, 651-669 (2013).
- S. Djebali, C.A. Davis, et al., Landscape of transcription in human cells. Nature. 489, 101-108 (2012).
- V. A. Moran, R. J. Perera, A. M. Khalil, Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 40, 6391 (2012).
- M. R. Fabian, N. Sonenberg, The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586-593 (2012).
- E. van Rooij and S. Kauppinen, Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 6, 851 (2014).
- F. Modarresi, M.A. Faghihi, N.S. Patel,et al., Knockdown of BACE1-AS Non-protein-Coding Transcript Modulates Beta-Amyloid-Related Hippocampal Neurogenesis. Int. J. Alzheimer's Dis. (2011), doi: 10.4061/2011/929042
- M.E. Askarian-Amiri, J. Crawford, J.D. French, et al., SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 17(5), 878-891 (2011).
- D. Khaitan, M.E. Dinger, J. Mazar, et al., The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 71(11), 3852-3862 (2011).
Order products
CRISPR Design Tool
Online design tool for CRISPR-Cas9 guide RNAs targeting microRNA or long noncoding RNA
Lincode siRNA
Predesigned human and mouse siRNA reagents, screening libraries, and validated controls for effective lncRNA knockdown
miRIDIAN microRNA Mimics and Hairpin Inhibitors
Synthetic RNA tools available as screening libraries or individual reagents
shMIMIC Inducible Lentiviral microRNA
The industry's most innovative mature microRNA expression system combined with latest generation of powerful Tet-inducible technology
shMIMIC Lentiviral microRNA
Expressed lentiviral microRNAs with 7 promoter and 2 reporter options for optimal, integrated expression of mature microRNAs in difficult-to-transfect cells
Helpful resources
Dharmacon microRNA Selection Guide
A quick tool to help determine the ideal Dharmacon microRNA product line and format for your needs
shMIMIC Inducible Lentiviral microRNA - Technical Manual
Advantages of the unique shMIMIC microRNA expression design with the Tet-On 3G tetracycline-inducible expression system
shMIMIC Lentiviral microRNA - Technical Manual
The Dharmacon SMARTchoice lentiviral vector platform is an innovative system ideally suited for RNA interference (RNAi)-mediated studies
