Power Your Immune-Oncology Projects with Custom Engineered Immune Cells
Gene-editing in immune cell lines can be challenging due to low targeting efficiency and difficulties in single cell derivation of suspension cells. Horizon has validated 10+ immune cell lines including THP-1, Jurkat and NALM-6 cells for gene-editing projects. CRISPR, rAAV and ZFN gene-editing technologies are available depending on project requirements.
Take advantage of the largest panel of pre-characterized immune cell lines available and benefit from Horizon’s exceptional knowhow and experience in completing over 2,000 gene-editing projects.
Gene-Editing for Immuno-Oncology Offering
- Modifications: Knockin, knockout (point mutations, tags and reporter genes)
- Technologies: CRISPR, rAAV or ZFN depending on project requirements
- Project types: Standard or Premium
- Estimated timeline: 13-32 weeks
- Deliverables: Isogenic cell line pair and a summary report
Cell Lines Including
|
Cell line |
Species |
Cell Type |
Disease |
|
TF-1 |
Human |
Erythroblast |
Erythroleukemia |
|
THP-1 |
Human |
Monocyte |
Acute monocytic leukemia |
|
MOLT-4 |
Human |
T-lymphoblast |
Acute lymphoblastic leukemia |
|
NALM-6 |
Human |
B-cell |
Acute lymphoblastic leukemia |
|
KMS-11 |
Human |
B-cell precursor |
Myeloma |
|
K-562 |
Human |
Lymphoblast |
Chronic myelogenous leukemia |
|
Jurkat |
Human |
T-lymphocyte |
Acute T-cell leukemia |
|
J.RT3-T3.5 |
Human |
T-lymphocyte |
Acute T-cell leukemia |
|
H9 |
Human |
Cutaneous T-lymphocyte |
Lymphoma |
|
CCRF-CEM |
Human |
T-lymphoblast |
Acute lymphoblastic leukemia |
|
A20 |
Mouse |
B-lymphocyte |
Reticulum cell sarcoma |
|
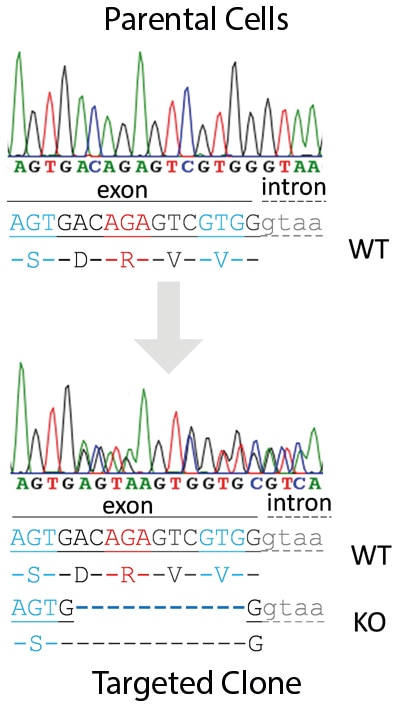
Case Study I: Heterozygous CRISPR Knockout in Jurkat Cells |
|
|
|
Positive clone with heterozygous knockout was identified by PCR and Sanger sequencing. An 11 bp deletion introducing a premature stop codon at position 51 exists in one allele. |
|
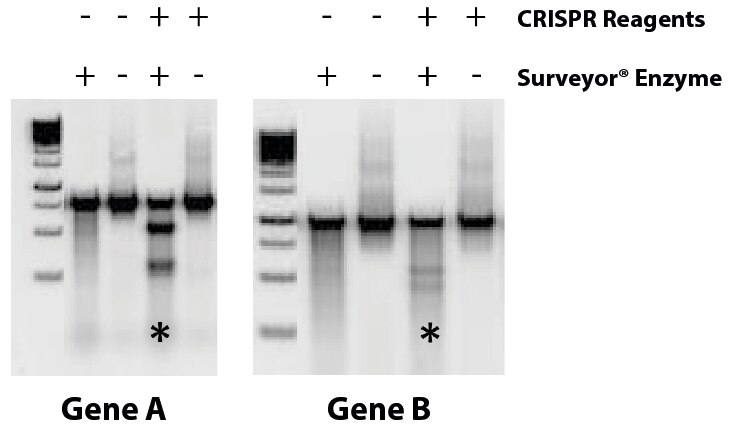
Case Study II: CRISPR-Mediated Gene Disruption in THP-1 Cells |
|
|
|
Following transfection of THP-1 cells with CRISPR targeting reagents directed against two genes, CRISPR mediated disruption of each gene was identified (indicated by *) by Surveyor assay. |