- Gene modulation
- RNA interference
- ON-TARGETplus Cyclophilin B Control Pool
ON-TARGETplus Cyclophilin B Control Pool

ON-TARGETplus Cyclophilin B Control siRNA is a validated positive control, guaranteed to silence cyclophilin B in human, mouse, or rat cells. The ON-TARGETplus siRNA patented dual-strand modification pattern reduces off-targets by up to 90%, allowing improved specificity.
Also known as peptidylprolyl isomerase B, cyclophilin B is associated with the secretory pathway. This gene is abundantly expressed in most cells and because it is non-essential, knockdown of the corresponding mRNA does not affect cell viability.
Highlights
- Ideal positive controls for RNAi experiments using human, mouse, or rat SMARTpool reagents
- Pool of four highly functional, validated siRNAs for guaranteed silencing of cyclophilin B in human, mouse, or rat cells
- Target accession numbers NM_000942 (human, D-001820-10), NM_011149 (mouse, D-001820-20), or NM_022536 (rat, D-001820-30)
Only the ON-TARGETplus modification pattern addresses both siRNA strands for premium specificity
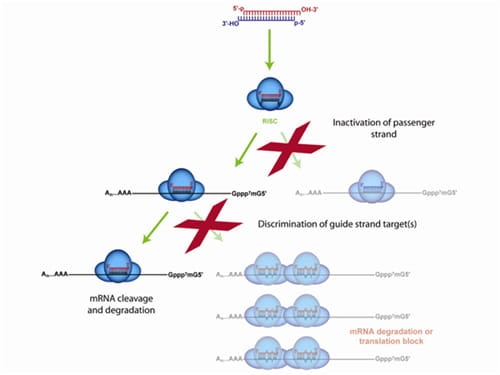
The ON-TARGETplus dual-strand chemical modification begins with the sense (passenger) strand being blocked from RISC uptake to favor antisense (guide) strand loading and reduce passenger strand-induced off-targets. However, the majority of siRNA off-targets are driven by the seed region of the guide strand. ON-TARGETplus is modified within its seed region to destabilize miRNA-like activity and improve specificity to the desired target for potent knockdown.
ON-TARGETplus modifications reduce the overall number of off-targets and pooling reduces them even further
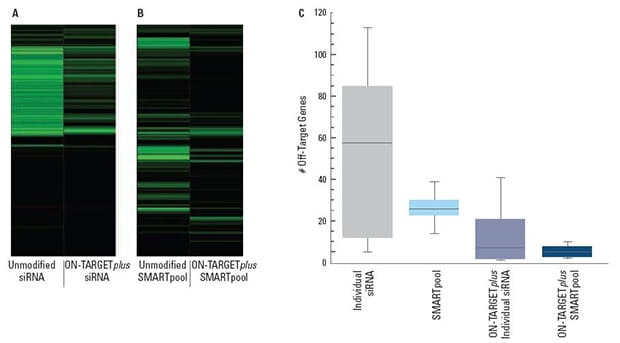
Panels (A) and (B) are representative examples of off-target signatures with and without application of ON-TARGETplus modifications to (A) a single siRNA and (B) a SMARTpool reagent. Green bars indicate genes with 2-fold or more reduction of expression when treated with the indicated siRNA reagent.The ON-TARGETplus modifications reduced the off-targets when compared to unmodified siRNA. Pooling of siRNA and the ON-TARGETplus modification pattern independently, and in combination, provide significant reduction in off-target gene silencing. Panel (C) represents quantitation of off-targets (down-regulated by 2-fold or more) induced by the indicated siRNA reagents targeting 10 different genes (4 siRNAs per gene or a single SMARTpool reagent). Off-targets were quantified using microarray analysis (Agilent) then compiled. Each shaded box represents the middle 50% of the data set. Horizontal line in box: Median value of the data set. Vertical bars: minimum and maximum data values.
