- Non-mammalian research tools
- Molecular Barcoded Yeast (MoBY) ORF Library
Molecular Barcoded Yeast (MoBY) ORF Library

The molecular barcoded yeast open reading frame (MoBY-ORF) library was initially created to provide insight into identifying drug resistance mutations in yeast. Through the use of MoBY-ORF complimentation cloning, spontaneous drug-resistant mutants can be identified via depletion of a unique molecular barcode. These barcodes permit use of a pooled screening strategy as well as simultaneous measurement of all transformants.
The MoBY-ORF 1.0 library was created by cloning each gene, controlled by its native promoter and terminator, into a centromere-based vector along with two unique oligonucleotide barcodes. The plasmid vector p5472 carries a URA3 selectable marker and a yeast centromere, and was designed to allow transfer of barcoded clones to other vector backbones through mating-assisted genetically integrated cloning (MAGIC).
The Charlie Boone lab at The University of Toronto has created the Molecular Barcoded Yeast ORF 2.0 (MoBY-ORF 2.0) library for drug-target screening applications in S.cerevisiae. Each gene from the MoBY-ORF 1.0 collection was transferred using MAGIC (Mating-Assisted Genetically Integrated Cloning) from the original centromere-based vector into a high-copy plasmid backbone (p5476), and transformed into the BY4741 yeast strain. Each gene was moved in its entirety and is controlled by its native promoter and terminator and has two unique DNA barcodes. The MoBYORF 2.0 resource has numerous genetic and chemical-genetic applications.Highlights
- The majority of MoBY-ORF clones contain only a single open reading frame. Normal expression patterns of ORFs are preserved through use of endogenous promoters and 3' UTRs
- Plasmids are barcoded by two unique 20-bp oligonucleotides, enabling measurement of the relative abundance of a specific transformant via barcode detection. Unique barcodes also permit simultaneous analysis of all transformants in a single pool
- Molecular barcodes comprise two unique 20-nucleotide DNA sequences (labeled the UPTAG and DNTAG), flanking a dominant selectable marker (KanMX) that confers resistance to the drug G418/kanamycin
- The MoBY-ORF 1.0 collection is delivered in E. coli; the MoBY-ORF 2.0 collection is delivered in S.cerevisiae
Molecular Barcoded Yeast (MoBY) ORFs Diagram
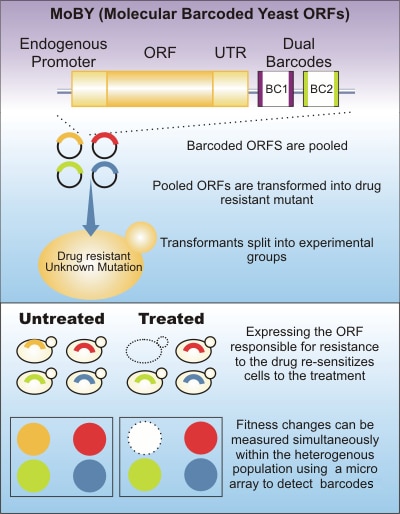
The use of molecular barcoded yeast ORFs can identify a recessive drug-resistant mutant by complementation cloning.
Applications
- Genome scale parallel screening
- Discover mechanism-of-action for drug discovery and mutation screening
Note
We provide certain clone resources developed by leading academic laboratories. Many of these resources address the needs of specialized research communities not served by other commercial entities. In order to provide these as a public resource, we depend on the contributing academic laboratories for quality control.
Therefore, these are distributed in the format provided by the contributing institution "as is" with no additional product validation or guarantee. We are not responsible for any errors or performance issues. Additional information can be found in the product manual as well as in associated published articles (if available). Alternatively, the source academic institution can be contacted directly for troubleshooting.
Clone Collections
The stock plates will be provided in 96-well microtiter plates. These will ship on dry-ice via overnight delivery and should be stored at –80°C immediately upon receipt. Please contact our customer service department for a quote and estimated shipping time on bulk orders.
- C. H. Ho et al., A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nature Biotechnology. 27(4), 369-377 (April 2009).
