- Non-mammalian research tools
- Yeast Synthetic Histone H3 and H4 Mutant Collection
Yeast Synthetic Histone H3 and H4 Mutant Collection

Nucleosome structural integrity underlies the regulation of DNA metabolism and transcription. Developed by Dr. Jef Boeke at Johns Hopkins University, the versatile library of 486 systematic histone H3 and H4 substitution, and deletion mutants was generated in Saccharomyces cerevisiae to investigate the contribution of each amino acid to nucleosome function. Each amino acid residue was systematically substituted with alanine, and all alanine residues were changed to serine.
The Yeast Histone H3 and H4 Mutant Collection is available in both yeast and bacteria. The bacterial collection contains all 486 mutants in Escherichia coli. In the yeast format, there is a non-essential and essential collection in a MATa yeast strain.
Highlights
- Contains 486 systematic histone H3 and H4 substitution and deletion mutants
- Each mutant was linked to two unique 20 bp barcode sequences, allowing for microarray-based analysis of the library for quantitative or phenotypic assays
- The yeast clones containing lethal mutations were transfected with an additional plasmid pJP11 (CEN LYS HHF1-HHT1) to compensate for the lethality and rearrayed into a separate collection, the Essential Histone H3 & H4 Mutant collection (Dai, 2008)
Note
We provide certain clone resources developed by leading academic laboratories. Many of these resources address the needs of specialized research communities not served by other commercial entities. In order to provide these as a public resource, we depend on the contributing academic laboratories for quality control.
Therefore, these are distributed in the format provided by the contributing institution "as is" with no additional product validation or guarantee. We are not responsible for any errors or performance issues. Additional information can be found in the product manual as well as in associated published articles (if available). Alternatively, the source academic institution can be contacted directly for troubleshooting.
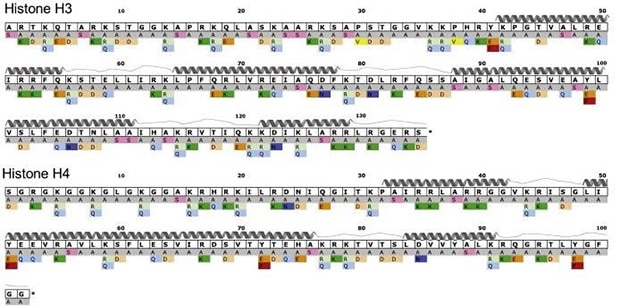
- J. Dai et al., Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 134(6), 1066-1078 (19 September 2008).
