- Non-mammalian research tools
- Yeast TAP Tagged ORFs
Yeast TAP Tagged ORFs

Yeast TAP Tagged ORFs
1Start Here
2Choose
The Yeast-TAP Tagged ORF library allows the purification and the selection of the yeast proteome and associated components using two affinity selection steps in tandem, enabling the development of a range of high-throughput functional assays. By tagging each ORF with a high-affinity epitope and expressing the protein from its natural chromosomal location, a census of proteins expressed during log-phase growth can be quantified through immunodetection of the common tag.
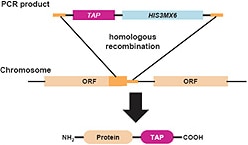
Yeast Tap Fusion
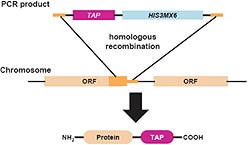
The C-terminal TAP insertion cassette contains the coding region for a modified version of the TAP (Tandem Affinity Purification) tag, which consists of a calmodulin binding peptide, a TEV cleavage site, two IgG binding domains of Staphylococcus aureus protein A, and a selectable marker.
Yeast Tap Fusion
The C-terminal TAP insertion cassette contains the coding region for a modified version of the TAP (Tandem Affinity Purification) tag, which consists of a calmodulin binding peptide, a TEV cleavage site, and two IgG binding domains of Staphylococcus aureus protein A as well as a selectable marker.
Highlights
- Allows for the purification and the selection of the yeast proteome and associated components using two simple affinity selection steps in tandem
- A membrane-bound protein subset has been arrayed in separate plates for your convenience
- Background strain = MatA (BY4741)
Note
We provide certain clone resources developed by leading academic laboratories. Many of these resources address the needs of specialized research communities not served by other commercial entities. In order to provide these as a public resource, we depend on the contributing academic laboratories for quality control.
Therefore, these are distributed in the format provided by the contributing institution "as is" with no additional product validation or guarantee. We are not responsible for any errors or performance issues. Additional information can be found in the product manual as well as in associated published articles (if available). Alternatively, the source academic institution can be contacted directly for troubleshooting.
Individual strains are provided in tubes containing glycerol stock cultures in YPD plus 15% glycerol and 100 µg/mL Carbenicillin. While the scTAP strains are supplied in YPD media, SD complete or SD -HIS media can also be used. Individual strains from this library can be queried and purchased through our clone query.
The TAP Library is provided in 96-well microtiter plates containing frozen glycerol stock cultures in YPD plus 15% glycerol and 100 µg/mL Carbenicillin. While the scTAP strains are supplied in YPD media, SD complete or SD-HIS media can also be used. Strains are arranged in plates by protein expression size from largest to smallest, with plate designation GS1 representing high expression and GS5 representing low expression. In addition, the membrane-protein subset has been arrayed in separate plates for your convenience. Refer to USB shipped with the collection for expression category designation.
The use of the TAP-tag for the purification of biomolecule/protein complexes is covered by granted patents in Europe and Australia (EP1105508B1, AU07629621), and patent applications in Canada, Japan, and US (see International Patent Publication No. WO-0009716).
For licensing information for use in purification of biomolecule/protein complexes, please contact Cellzome AG, Meyerhofstrasse 1, 69117 Heidelberg, Germany, at info@cellzome.com.
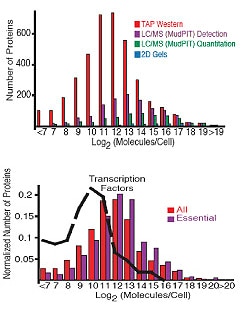
The abundance of proteins ranged from fewer than 50 to more than 106 molecules/cell with many, including essential proteins and most transcription factors, having levels not readily detectable by other proteomic techniques, nor predictable by mRNA levels or codon bias measurements. The abundance of proteins ranged from fewer than 50 to more than 106 molecules/cell with many, including essential proteins and most transcription factors, having levels not readily detectable by other proteomic techniques, nor predictable by mRNA levels or codon bias measurements.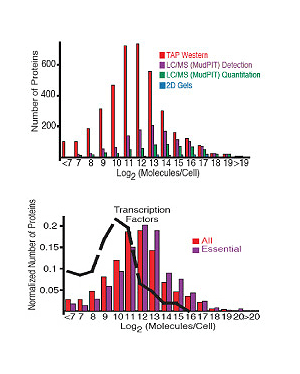
Yeast Tap Fusion - 1
Yeast Tap Fusion - 1
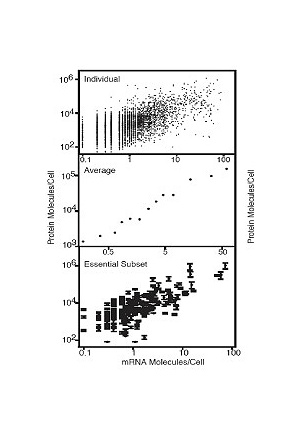
Yeast Tap Fusion - 2
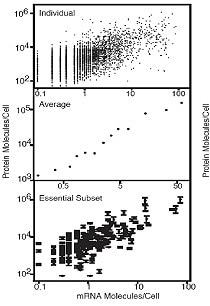
Yeast Tap Fusion - 2
- S. Ghaemmaghami et al., Global Analysis of Protein Expression in Yeast. Nature. 425(6959), 737-741 (16 October 2003). [Letters to Nature]
