- Dharmacon Screening libraries
- Edit-R Lentiviral sgRNA Pooled Screening Libraries
Edit-R Lentiviral sgRNA Pooled Screening Libraries
Ensure library specificity with algorithm-optimized guide RNA. Efficient gene editing at single-copy integrations.
Whole genome, druggable genome, gene family and pathway libraries for knockout screening.
Please Note: Human Pooled Libraries have been updated with a new and improved vector scaffold. Please reach out to Scientific Support if you have any questions.

Perform unbiased, phenotypic knockout screens without the need for costly infrastructure
Knockout hundreds and even thousands of genes at the DNA level through pooled lentiviral library screening methods.
The ability of CRISPR guide RNA to create a double-strand break in target DNA causing functional protein disruption can vary based on the guide RNA (gRNA) sequence and position in the targeted gene. To address this, the Dharmacon reagents algorithm provides highly specific and functional RNA guides across the human genome and mouse genomes.
The Edit-R CRISPR RNA algorithm generates guide RNA sequences with:
- An optimized alignment program with rapid and thorough specificity analysis, including detection of multiple mismatches and gapped alignments
- Increased functional knockout of targeted genes through systematic assessment of over 1100 guide RNAs to identify characteristics critical for functional gene disruption, not just generation of indels
- New! Edit-R human sgRNA designs have been updated to the latest RefSeq in 2025 providing the most specific and genomically relevant guides for producing efficient protein knockout. This allows the Edit-R algorithm to target the latest genome annotations more accurately and efficiently providing you with the best solution for your research needs. Please reach out to Scientific Support if you have any questions.
Highlights:
- Efficient two-vector system utilizing a lentiviral vector for Cas9 expression and a gene-specific vector for sgRNA expression
- Rationally designed Edit-R lentiviral sgRNAs efficient gene knockout with unparalleled specificity using a functionally validated proprietary algorithm
- Deep and broad coverage of 4 or 8 sgRNAs per gene across the human genome for whole genome library or 5 -10 sgRNAs per gene across the mouse genome for increased hit confidence.
- Can be combined with the promoter flexibility of Edit-R Lentiviral Cas9 Nuclease Reagents for robust editing in biologically relevant cell types
The NEW Edit-R Human Lentiviral sgRNA Pooled Screening Libraries include, per pool:
- ≥ 5 x 108 TU/mL ( ± 20%) lentiviral particles in 10 x 100 uL pack size
- Four or eight sgRNAs per gene (whole genome) and up to four sgRNAs per gene (gene family and pathway libraries) targeting coding genes in the NCBI Reference Sequence Database
- 100 non-targeting sgRNA negative controls bioinformatically confirmed to not align with (target) any gene in the human genome
- Gene family and pathway libraries include up to 136 gene-specific sgRNA technical controls targeting up to 34 protein-coding genes (4 sgRNA per gene); including common reference genes (ACTB, GAPDH, LAMB1, & PPIB)
- A data file containing complete library information, including: gene annotations, sgRNA target sequences, complete list of controls, and counts per millions of mapped reads
The Edit-R Mouse Lentiviral sgRNA Pooled Screening Libraries include, per pool:
- ≥ 5 x 108 TU/mL ( ± 20%) lentiviral particles in pre-aliquoted tubes
- 8 x 25 µL (200 µL total) for pools ≤ 5000 constructs
- 16 x 25 µL (400 µL total) for pools > 5000 constructs
- Five to ten sgRNAs per gene targeting coding genes in the NCBI Reference Sequence Database
- 100 non-targeting sgRNA negative controls bioinformatically confirmed to not align with (target) any gene in the human and mouse genomes
- Up to 340 gene-specific sgRNA positive controls targeting up to 34 protein-coding genes (10 sgRNA per gene); including common reference genes (ACTB, GAPDH, LAMB1, & PPIB)
- A data file containing complete library information, including: gene annotations, sgRNA target sequences, complete list of controls, and counts per millions of mapped reads
Products recommended in our validated protocol:
- Vector-matched Edit-R lentiviral positive and non-targeting sgRNA controls for gene editing validation and experimental optimization
- SMARTvector Non-targeting Control Particles with optimal promoter and fluorescent reporter (TurboGFP or TurboRFP) for determining relative transduction efficiency and optimizing transduction conditions in desired cells of interest.
- Edit-R Lentiviral Cas9 Nuclease Reagents or Cas9 Stable Cell Line
- Human libraries in NEW V2 backbone:
- NEW: NGS Library Prep Kit (Recommended)
- Primer set options available two different sizes for 12 or 24 whole genome screens
- Designed to amplify and sequence sgRNA constructs from gDNA without bias
- One complete solution for sequencing on an Illumina® platform without the need for additional reagents
- Mouse libraries and Human libraries in V1 backbone:
- Edit-R Mouse Pooled sgRNA Indexing PCR and Sequencing Primer Kit A for Illumina® sequencing platforms (required)
- Edit-R Mouse Pooled sgRNA Indexing PCR and Sequencing Primer Kit B for additional reverse indexing PCR primers (if experimental design requires additional multiplexing capability)
Important Notice
The Edit-R Lentiviral sgRNA Pooled Libraries are solely for internal research use (as set forth in the Product Terms and Conditions) in laboratories where the containment measures stated below and in applicable laws and regulations are met. Products may not be used for diagnostic, therapeutic or other commercial purposes and may not to be administered to humans for any purpose or to animals for therapeutic purposes. Edit-R Lentiviral sgRNA Pooled Libraries provided as lentiviral particles are replication-incompetent, self-inactivating (SIN) and non-pathogenic (do not cause infectious human disease).
Any investigator who purchases lentiviral particle products is responsible for consulting with their institution's health and biosafety personnel for specific guidelines on the handling of lentiviral vector particles. Furthermore, each investigator is fully responsible for obtaining the required permissions for research using and the acceptance of replication-incompetent SIN lentiviral vectors and replication-defective lentiviral particles into their local jurisdiction and institution.
** Please contact Scientific Support for more information
Approximate number of targeted genes in respective human or mouse Edit-R Lentiviral sgRNA pooled libraries. Please reach out to Scientific Support for more information regarding gene composition of each library.
| Approximate Number of targeted genes | ||
|---|---|---|
| Human | Mouse | |
| Whole Genome | 19,000 | 19,680 |
| Druggable Genome | 8,350 | 9,930 |
| GPCRs | 480 | 510 |
| Ion Channels | 430 | 340 |
| Protein Kinases | 760 | 780 |
| Phosphatases | 310 | 270 |
| Proteases | 700 | 700 |
| Ubiquitin Conjugate | 660 | 660 |
| Epigenetics | 860 | 770 |
| Cytokine Receptors | 140 | 140 |
| Membrane Trafficking | 140 | 110 |
| Deubiquitinating Enzymes | 110 | 100 |
| Cell Cycle Regulation | 170 | 120 |
| Tyrosine Kinases | 90 | 90 |
| Nuclear Receptors | 50 | 50 |
| Apoptosis | 550 | 290 |
| DNA Damage Response | 240 | |
Volume of lentiviral particles required for screening with Edit-R Human Whole Genome library
| Fold representation | Replicates | sgRNAs per gene | Volume of lentiviral particles |
|---|---|---|---|
| 200 | 2 | 4 | 0.07 mL |
| 200 | 2 | 8 | 0.13 mL |
| 400 | 2 | 4 | 0.13 mL |
| 400 | 2 | 8 | 0.25 mL |
| NGS Library Prep Kit Contents | Small: 12 Whole Genome Screens | Large: 24 Whole Genome Screens |
|---|---|---|
| NEXTFLEX® PCR Master Mix (green cap) | 3600 μL | 7200 μL |
| Dharmacon HIT Identification Primers (orange cap) | 300 μL | 600 μL |
| NEXTFLEX® PCR II Barcoded Primer Mix | 4 μL | 4 μL |
| Resuspension Buffer | 12 mL | 24 mL |
| Nuclease-free water | 8 mL | 16 mL |
| Cleanup Beads | 7 mL | 15 mL |
Gene knockout screening workflow using the Edit-R Lentiviral sgRNA Pooled Screening Library platform
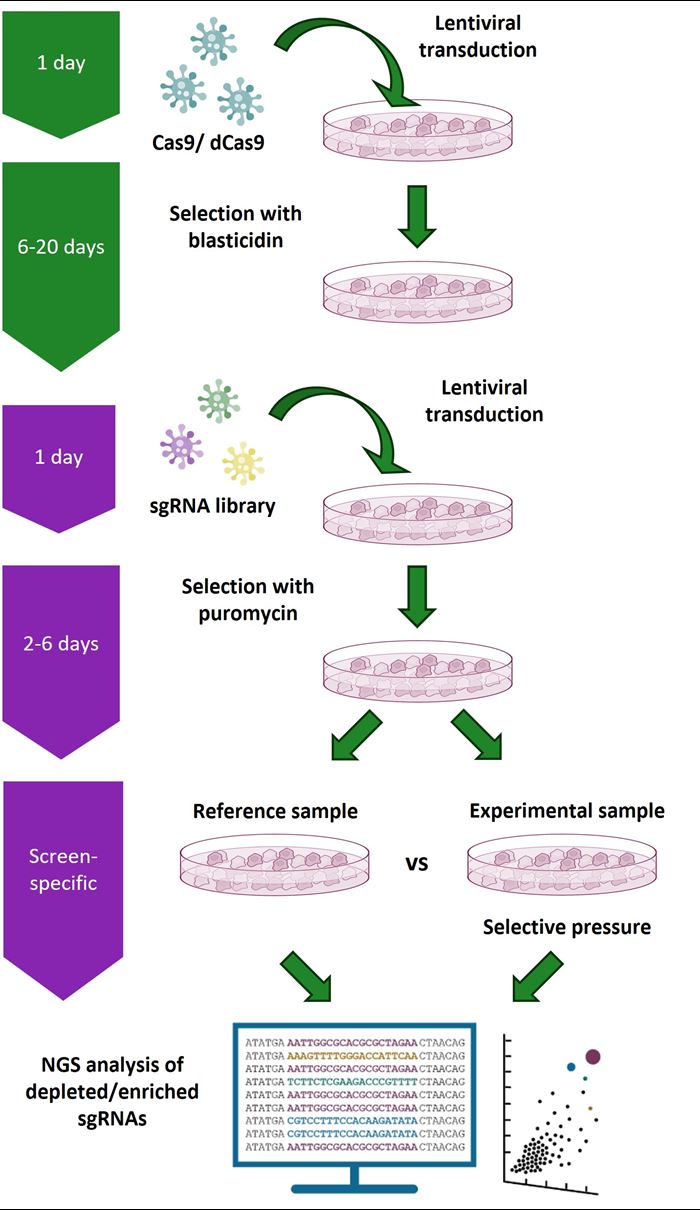
A Cas9-expressing stable cell line (mixed or clonal cell population) is first generated with Edit-R Lentiviral Cas9 Nuclease particles by selection with blasticidin. These cells are then transduced with an Edit-R lentiviral sgRNA pooled library and selected with puromycin. Transduced cells are split into reference and experimental populations for application of a selective pressure and/or phenotypic selection. Genomic DNA is then isolated from the reference and experimental populations of transduced cells and sgRNA constructs within the isolated gDNA are amplified vector-specific NGS Library Prep Kits. Samples can then be multiplexed and sequenced on an Illumina® platform. Custom sequencing read primers are not required. The integrated sgRNA sequences in both reference and experimental samples are identified and relative abundance compared. sgRNA constructs that are enriched or depleted are identified as hits to be confirmed and studied further using individual Edit-R Lentiviral sgRNAs in additional phenotypic and/or biochemical assays.
Schematic maps of the Edit-R Lentiviral Cas9 Nuclease and sgRNA vectors
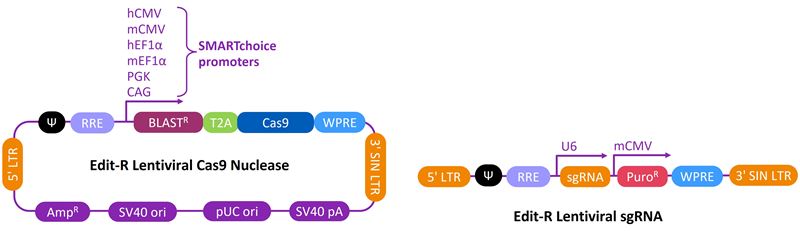
Schematic maps and table of vector elements of the Edit-R Lentiviral Cas9 Nuclease (A) and sgRNA (B) vectors. The Edit-R Lentiviral CRISPR-Cas9 platform is a two-vector system that utilizes a lentiviral vector with multiple Pol II promoter options for maximal Cas9 expression and a gene-specific vector for sgRNA expression designed to the target site of interest.
High quality pooled screening begins with rigorous lentiviral sgRNA pooled library production
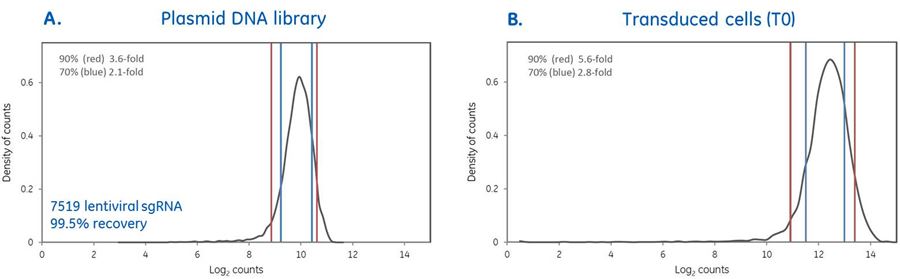
High quality pooled screening begins with rigorous lentiviral sgRNA pooled library production. A pooled library comprised of 7519 lentiviral sgRNAs targeting human kinases was produced and the quality of the plasmid DNA library was verified by next-generation sequencing (NGS). Counts per million mapped reads were obtained to determine percent-recovery of input sgRNAs (99.5%) and the distribution of 90% and 70% of the sgRNAs in the pool are within 3.6- and 2.1- fold of each other, respectively (Panel A.). The pooled plasmid DNA library was then packaged into lentiviral particles and transduced at MOI 0.3 into U2OS cells from which genomic DNA was isolated at 24 hours post-transduction (T0), PCR-amplified and prepped for NGS. Distribution of lentiviral sgRNAs was not substantially affected, indicating rigorous production and maximized retention of constructs from production into pooled screening (Panel B.).
Pooled lentiviral sgRNA human kinase library screen identifies high-confidence viability hits
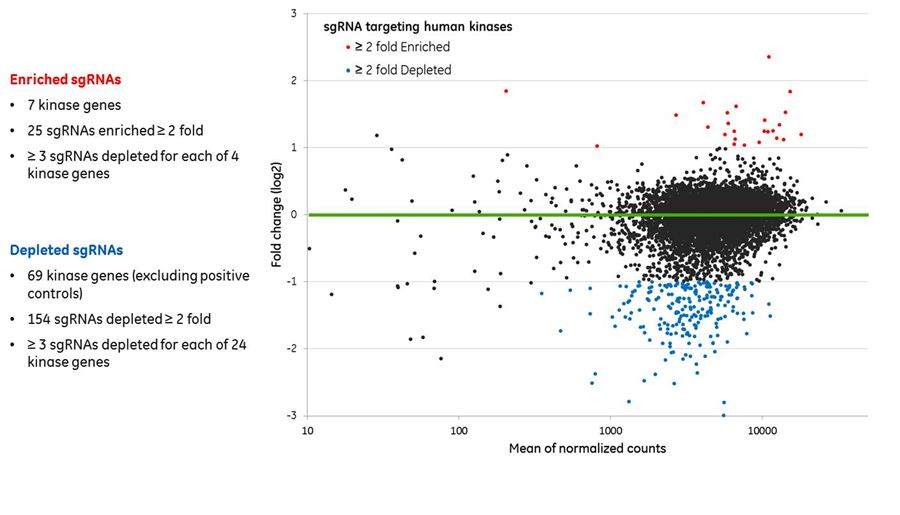
Pooled lentiviral sgRNA human kinase library screen identifies high-confidence viability hits. A pooled lentiviral library comprised of 7079 Edit-R sgRNAs targeting 708 human kinases, 340 positive control sgRNAs targeting 34 essential genes and 100 non-targeting control sgRNAs was transduced into a Cas9-expressing U2OS cell line at MOI 0.3 and 1000-fold sgRNA representation with two biological replicates. At 24 hours post-transduction, the reference (T0) cell populations were harvested and the experimental (T1) cell populations were subjected to 2 ug/mL puromycin treatment (selective pressure). At 8 days post-selection T1 cell populations were harvested. Genomic DNA was harvested from both T0 and T1 samples and PCR-amplified for high-throughput sequencing on an Illumina platform. sgRNA abundance is shown as an MA plot of normalized count data. The sgRNAs that were significantly (adjusted p-value ≤ 0.05) higher and lower abundance and also showed > 2-fold abundance change are in red and blue, respectively. The green line indicates zero fold abundance change.
