- Dharmacon Screening libraries
- Edit-R synthetic sgRNA libraries
Edit-R synthetic sgRNA libraries
Simplify CRISPR editing in all cell types including primary cells with Edit-R synthetic sgRNA libraries
Ensure screen hit specificity - algorithm optimized synthetic single guide RNA.
Arrayed collections of pre-designed synthetic sgRNA for screening across the human genome and gene families. Use in rapid loss-of-function studies.

Further your functional genomics studies with CRISPR-Cas9 gene editing using Edit-RTM synthetic sgRNA libraries. Analyze hundreds of genes with multiple target sites per gene. Collections include whole genome, human druggable genome, and common gene family libraries
Arrayed library format permits single-well analysis with many phenotypic assays; including high-content assays and other morphological or reporter assays.
New! Edit-R human sgRNA designs have been updated to the latest RefSeq in 2025 providing the most specific and genomically relevant guides for producing efficient protein knockout. This allows the Edit-R algorithm to target the latest genome annotations more accurately and efficiently providing you with the best solution for your research needs. Please reach out to Scientific Support if you have any questions.
Highlights of Edit-RTM synthetic sgRNA libraries:
- Selected by the Edit-RTM algorithm. Highly functional and specific target sequences for robust, reliable gene knockout. Chemically modified for improved nuclease resistance
- For high-confidence phenotypic results, provided as three unique sgRNA designs per gene, as a pooled reagent
- Conveniently arrayed in 96-or 384-well plates and offered as gene family collections. Echo-qualified 384-well plates are available upon request
- Also available as sgRNA cherry-pick libraries; simply upload your own gene list and customize your plate
Human Druggable is made up of: Proteases, Protein Kinases, Phosphatases, Transcription Factors, Ubiquitin Enzymes, GPCRs, Ion Channels and Drug Targets.
| Human synthetic sgRNA libraries | Approximate number of genes |
|---|---|
| Whole Genome | 18,880 |
| Druggable Genome | 8,270 |
| GPCRs | 470 |
| Ion Channels | 430 |
| Phosphatases | 310 |
| Proteases | 700 |
| Protein Kinases | 760 |
| Ubiquitin Enzymes | 750 |
| Transcription Factors | 1580 |
| Drug Targets | 3300 |
| Apoptosis | 550 |
| Cell Cycle Regulation | 170 |
| Cytokine Receptors | 140 |
| Deubiquitinating Enzymes | 110 |
| DNA Damage Response | 240 |
| Epigenetics | 860 |
| Membrane Trafficking | 140 |
| Nuclear Receptors | 50 |
| Tyrosine Kinases | 90 |
Flow cytometry analysis of protein KO in CD4+ T cells with a pool of predesigned synthetic sgRNA.
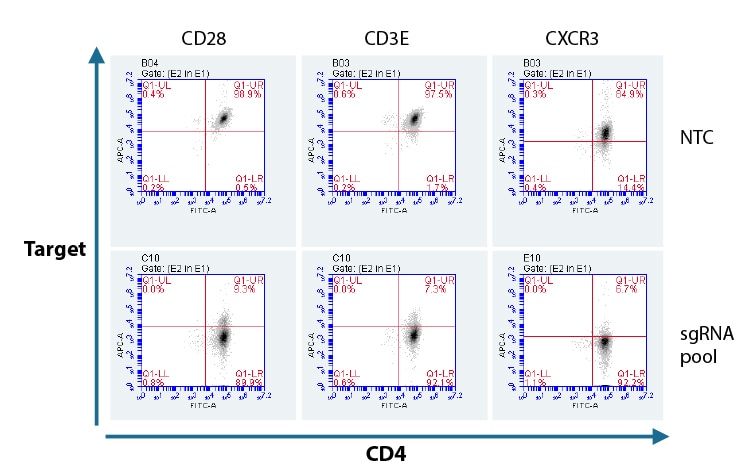
Primary human CD4+ T cells were nucleofected with Cas9 RNP using a pool of three Edit-R predesigned synthetic sgRNAs, or a Non-targeting control (NTC) via a Lonza 96-well Shuttle system. After 72 hours, functional knockout of CD28, CD3E, and CXCR3 were assessed as a percent of cells not expressing the target gene by FACS analysis. Cells were stained for CD4 as a positive expression control using an Alexa Fluor 488 conjugated antibody and compared to each targeted gene knockout CD28, CD3E, and CXCR3 using APC conjugated primary antibodies.
Comparison of synthetic guide RNA formats in gene knockout in CD4+ T cells.
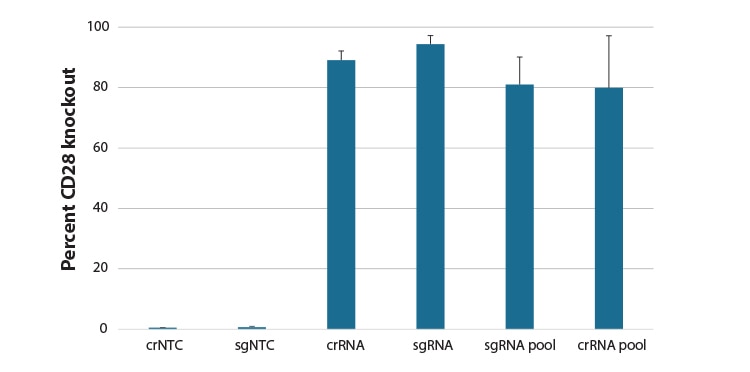
Primary human CD4+ T cells were nucleofected with a Cas9 RNP using an Edit-R predesigned synthetic crRNA:tracrRNA, synthetic sgRNA, pool of 3 different predesigned synthetic sgRNA, or pool of 4 predesigned crRNA:tracrRNAs targeting CD28, or a Non-targeting control (NTC) via a Lonza 96-well Shuttle system. After 72 hours, functional knockout of CD28 was assessed as a percent of CD28 negative cells by FACS analysis using an APC conjugated primary antibody to CD28.
Functional knockout of FUS gene with a pool of synthetic sgRNA.
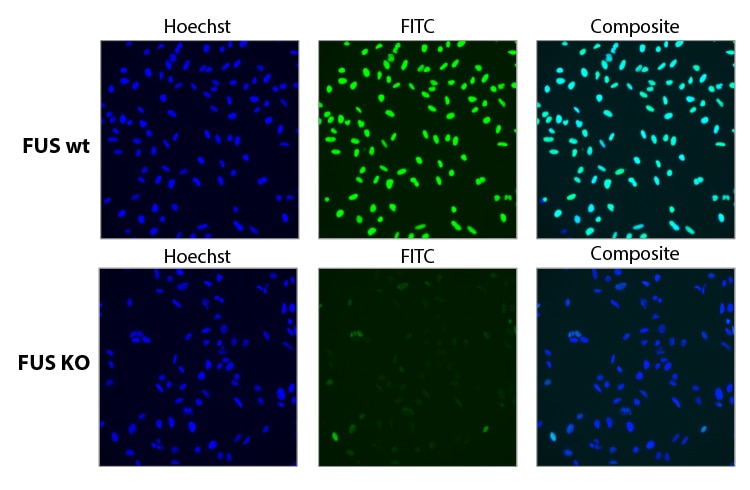
Functional knockout of FUS was assessed in U2OS cells constitutively expressing Cas9 under the CAG promoter. Cells were transfected with a 25 nM sgRNA pool containing three different Edit-R predesigned sgRNAs targeting FUS using 0.04 µL DharmaFECT 4. 72 hours post transfection the cells were split, and at 96 hours post transfection, cells were fixed and stained with a primary antibody targeting FUS, and an Alexa Fluor 488 conjugated fluorescent secondary antibody. Hoechst stain was used to identify nuclei.
Nuclease stabilization modifications improve gene editing efficiency
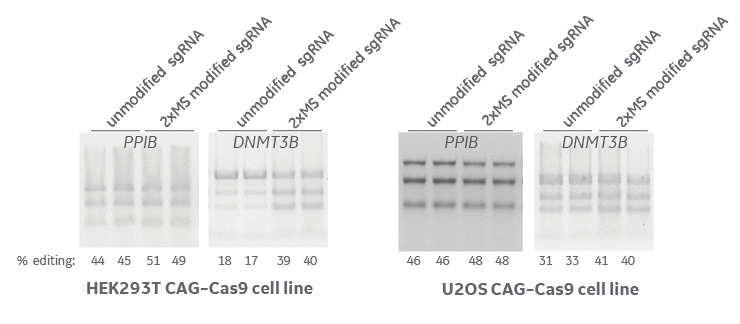
HEK293T and U2OS integrated Cas9 (under the CAG promoter) cell lines were transfected with unmodified and 5’ and 3’ 2xMS modified synthetic sgRNA targeting PPIB and DNMT3B genes. At 72 hours editing efficiency was assessed with a mismatch detection assay, T7 Endonuclease I (T7EI). In all cases the stabilizing modifications improve gene editing efficiency. MS = 2’-O-methyl nucleotides and phosphorothioate backbone linkage.
Edit-R synthetic guide RNAs cause virtually no innate immune response or toxicity compared to in vitro transcribed guide RNA

A HEK293T Cas9 nuclease expressing cell line was transfected with different synthetic guide RNA formats including unmodified crRNA:tracrRNA, crRNA:tracrRNA modified with 2xMS on 5’ crRNA and 3’ tracrRNA, crRNA:tracrRNA modified with 3xMS on both 5’ and 3’ crRNA and tracrRNA, unmodified synthetic sgRNA, modified synthetic sgRNA with 5’ and 3’ 2xMS or 3xMS, and in vitro transcribed (IVT) sgRNA targeting PPIB and DNMT3B genes. At 72 hours viability was assessed with the Resazurin reduction assay (red dots) and the levels of five immune response genes were quantified by RT-qPCR. MS = 2’-O-methyl nucleotides and phosphorothioate backbone linkages.
Structure of modifications for improved nuclease resistance on Edit-R synthetic sgRNA
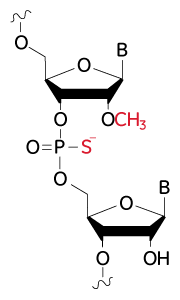
Modifications for improved nuclease resistance: 2’-O-methyl modified nucleotides and phosphorothioate backbone linkages.
Sequence structure of Edit-R synthetic single guide RNA
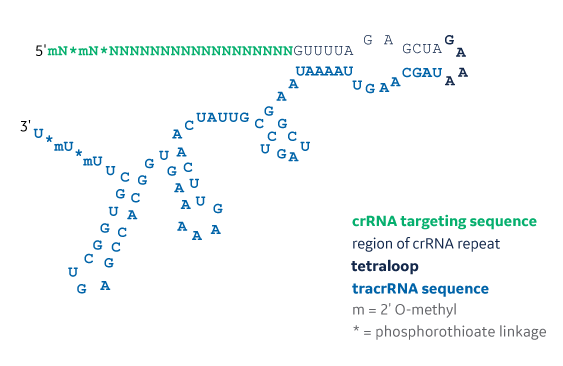
The structure of a Edit-R synthetic single guide RNA showing the 2xMS nuclease stability modifications on both 5’ and 3’ ends of the molecule. The structure contains a 20 nt targeting sequence (shown in green as poly Ns), 12 nt crRNA repeat sequence (shown in light gray), 4 nt tetraloop sequence (shown in bold black), and a 64 nt tracrRNA sequence (shown in blue). MS = 2’O-methyl nucleotides and phosphorothioate linkages (MS).
- R. Barrangou, A. Birmingham, et. al. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Research, 43(7) 3407-3419 (2015)
- M.L. Kelley, Ž. Strezoska, et al. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J. Biotechnol. 233, 74–83 (2016). doi:10.1016/j.jbiotec.2016.06.011
- Basila, M., M. L. Kelley, et al.. Minimal 2’-O-methyl phosphorothioate linkage modification pattern of synthetic guide RNAs for increased stability and efficient CRISPR-Cas9 gene editing avoiding cellular toxicity. PLoS One. 12, e0188593 (2017). doi: 10.1371/journal.pone.0188593
- Anderson, E.M., A. Haupt, et al. Systematic analysis of CRISPR-Cas9 mismatch tolerance reveals low levels of off-target activity. J. Biotechnol. 211, 56-65 (2015)
- He, K., E. Chou, et al. Conjugation and evaluation of triazole-linked single guide RNA for CRISPR-Cas9 gene editing. ChemBioChem. DOI: 10.1002/cbic.201600320 (2016)
