- Dharmacon Screening libraries
- Human Accell siRNA Library - Epigenetics
Human Accell siRNA Library - Epigenetics
Targeted gene silencing in difficult-to-transfect cells

The Accell Epigenetics library targets genes which encode histone and DNA modification factors like methyltransferases, acetyltransferases, kinases and deacetylases, in addition to genes with a role in chromatin remodeling and histone ubiquitination.
Accell siRNA achieves what no other RNAi product can claim: delivery into difficult-to-transfect cells without transfection reagents, virus, or electroporation. Researchers around the world are achieving targeted gene silencing in cells that had previously been beyond the reach of conventional RNAi products due to toxicity caused by transfection reagents or undesirable viral responses.
Highlights
- Accell siRNA enters cells without the need for transfection reagents, virus (or viral vectors), or instruments
- Proven performance in neuronal, immunological, primary, and other difficult-to-transfect cell types
- Extended-duration knockdown with optimized continuous application
- Available as SMARTpool siRNA reagents or a Set of 4 siRNAs in 96 or 384-well plates
Gene Targets
For a complete list of target genes in this siRNA Library, please contact Scientific Support.
Experimental Considerations
- Accell siRNA works at a higher concentration than conventional siRNA; recommended 1 µM working concentration
- Delivery may be inhibited by the presence of BSA in serum. Optimization studies with serum-free media formulations (Accell Delivery Media) or < 2.5% serum in standard media is recommended
- Full-serum media can be added back after 48 hours of incubation. Optimal mRNA silencing is typically achieved by 72 hours or up to 96 hours for protein knockdown
Cell types demonstrating effective silencing with Accell siRNA
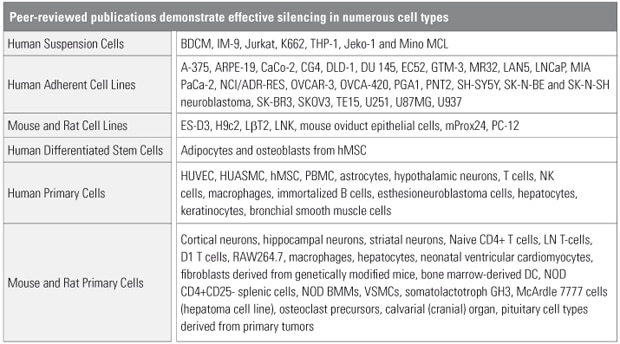
Cell types demonstrating effective silencing with Accell siRNA. Internal validation and peer-reviewed publications report numerous successes with difficult-to-transfect cell types. See the References tab for a list of publications.
The Accell siRNA application protocol simplifies targeted gene knockdown
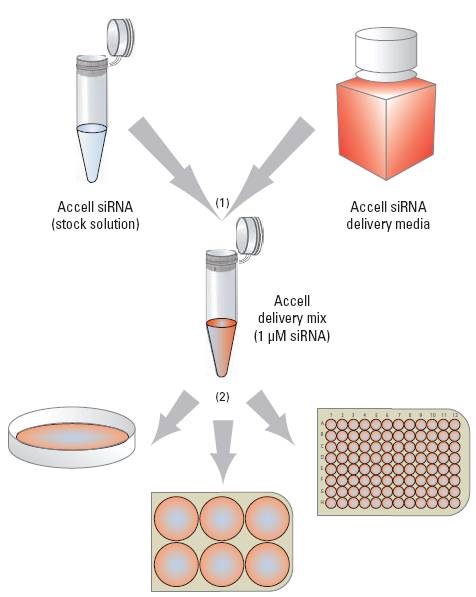
(A) Combine Accell siRNA with Accell delivery media (or other low- or no-serum media). (B) Add Accell delivery mix directly to cells, and incubate for 72 hours.
Click to view a more comprehensive list of Accell siRNA citations.
- B. D. Parsons, A. Schindler, D. H. Evans, E. Foley, A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS One. 4(12), e8471 (2009).
- M. Jiang, R. Instrell, B. Saunders, H. Berven, M. Howell, Tales from an academic RNAi screening facility; FAQs. Brief Funct. Genomics. 10(4), 227-237 (2011). [doi: 10.1093/bfgp/elr016]
- S. Suzuki et al., Differential Roles of Epac in Regulating Cell Death in Neuronal and Myocardial Cells. J. Biol. Chem. 285, 24248-24259 (July 2010). [primary mouse cortical neurons (E15-17)]
- U. Dreses-Werringloer et al., A Polymorphism in CALHM1 Influences Ca2+ Homeostasis, Ab Levels, and Alzheimer’s Disease Risk. Cell. 133, 1149-1161 (27 June 2008). [SHSY-5Y; human neuroblastoma]
- S. Byas et al., Human Embryonic Stem Cells Maintain Pluripotency after E-Cadherin Expression Knockdown. FASEB J. 24, lb172 (Apr 2010). [H9 stem cell lines]
- V. Saini et al, CXC Chemokine Receptor 4 Is a Cell Surface Receptor for Extracellular Ubiquitin. J. Biol. Chem. 285, 15566-15576 (May 2010). [THP-1 monocytes]
- D. Smirnov et al., Genetic Analysis of Radiation-induced Changes in Human Gene Expression. Nature. 459, 10.1038/nature07940(28 May 2009). [immortalized B cells]
