Parkinson's Disease (PD) is a complex neurodegenerative disorder that affects nearly 1 in every 500 people and can be either idiopathic, acquired or hereditary.
Genetic research has identified six distinct chromosomal regions containing genes with mutations that conclusively cause monogenic PD, where a mutation in a single gene is sufficient to cause the phenotype.
Parkinson's Disease (PD) is a complex neurodegenerative disorder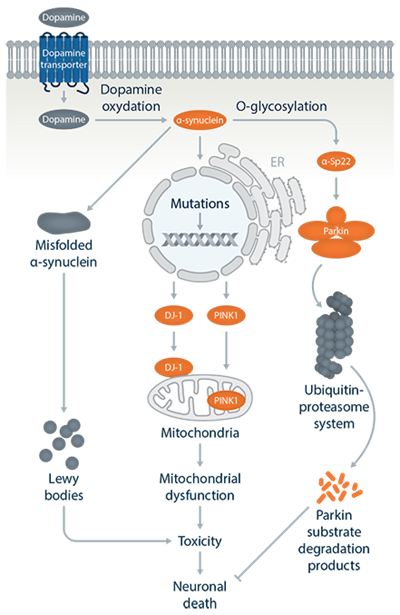
Pathways Parkinsons Disease
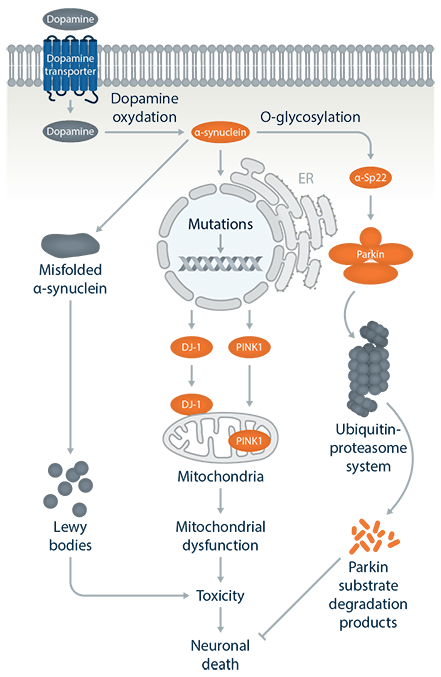
Pathways Parkinsons Disease
Pathogenic mechanisms
- The SNCA gene encodes an abundant 140-amino acid cytosolic protein, a-synuclein. Missense SNCA mutations cause PD through a toxic functional enhancement.
- LRRK2 gene mutations are the most frequent known cause of late-onset autosomal dominant and sporadic PD, but the pathogenic mechanisms are still unclear.
- PARK2 is the second largest gene in human genome and the 465-amino acid Parkin protein functions as an E3 ubiquitin ligase in the process of ubiquitination.
- PINK1 encodes a ubiquitously expressed protein kinase, which targets mitochondria. The majority of reported mutations in PINK1 are loss-of-function mutations affecting the kinase domain.
- PARK7 is ubiquitously expressed and functions as a cellular sensor of oxidative stress, but mutations linked to PD show reduced neuro-protective function and antioxidant activity.
- The ATP13A2 protein is a lysosomal membrane protein, and most mutations are truncated and unstable and so are retained and degraded in the endoplasmic reticulum.
In addition to these, several genes may be new targets for treatment by either turning on (TOMM7 and HSPA1L) or off (BAG4) gene activity.
Explore our available knockouts for Parkinson's disease
| SNCA | PINK1 | ATP13A2 | HSPA1L |
| LRRK2 | PARK7 | TOMM7 | BAG4 |
| PRKN |
Order products
Human knockout HAP-1 cells
The single largest bank of isogenic cell lines with over 7,500 cell lines to choose from and trusted by academia, biotech, and pharma research labs.
Cancer-related cell lines
Choose from over 300 knock-in and knockout cell line models in many standard cancer cell lines such as DLD1, MCF10A, and HCT116.
Cas9 Stable Cell Lines
Simplify gene editing experiments with stably expressing Cas9 cell lines
CRISPRmod CRISPRa dCas9-VPR Stable Cell Lines
Streamline CRISPR activation experiments with stably expressing dCas9-VPR cell lines
