The MAD7 for Gene Editing Design Tool allows you to quickly and easily generate guide RNA sequences for MAD7 and other CRISPR-Cas systems such as Cas12a, Cas12c, Cas13, or novel CRISPR enzymes.
Explore our products for successful CRISPR-Cas genome engineering
-
Edit-R CRISPR design tool
Our CRISPR Design Tool provides an intuitive one-stop location for guide RNA design and ordering
CRISPR genome engineering
Genome engineering has advanced tremendously with the characterization of bacterial and archaeal CRISPR (clustered regularly interspaced short palindromic repeats) systems and their adapted usage in mammalian cells. The ability to precisely and permanently alter endogenous genomic sequence through targeted genome editing is a highly effective genetics tool.
In addition to the canonical Cas9 nuclease, other editing nucleases have been discovered including Cas12a (includes MAD7), Cas12b, Cas12c, Cas13, and many more.
Gene editing with MAD7 nuclease
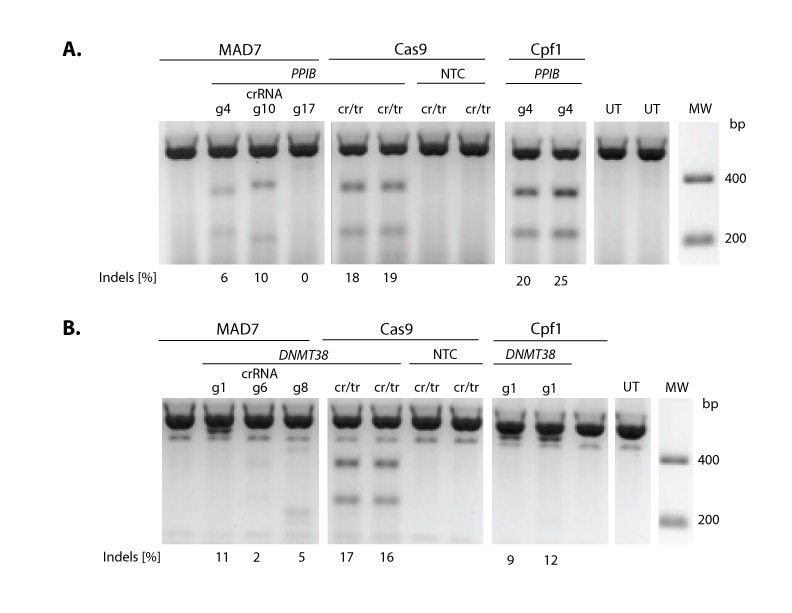
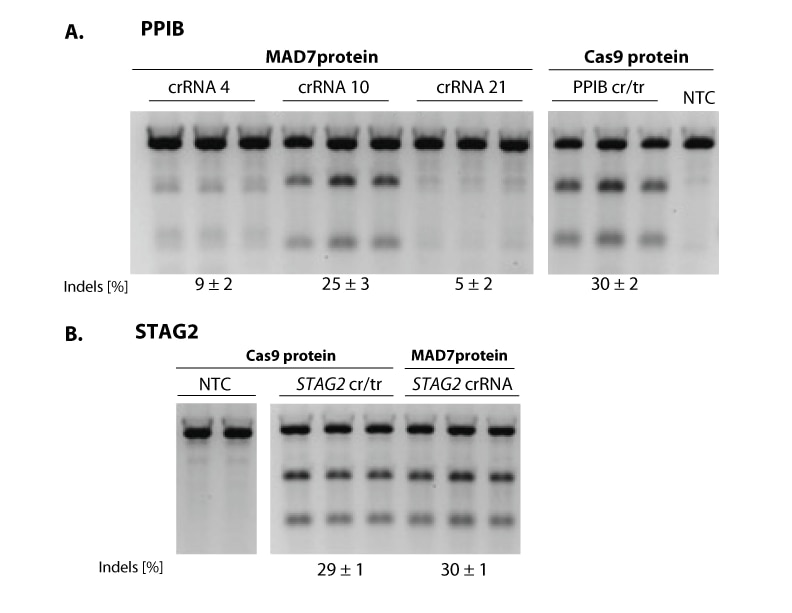
MAD7 is a novel nuclease that was released by Inscripta in 2017. This nuclease only requires a crRNA for gene editing and allows for specific targeting of AT rich regions of the genome. MAD7 cleaves DNA with a staggered cut as compared to S. pyogenes which has blunt cutting.
Gene Editing with MAD7
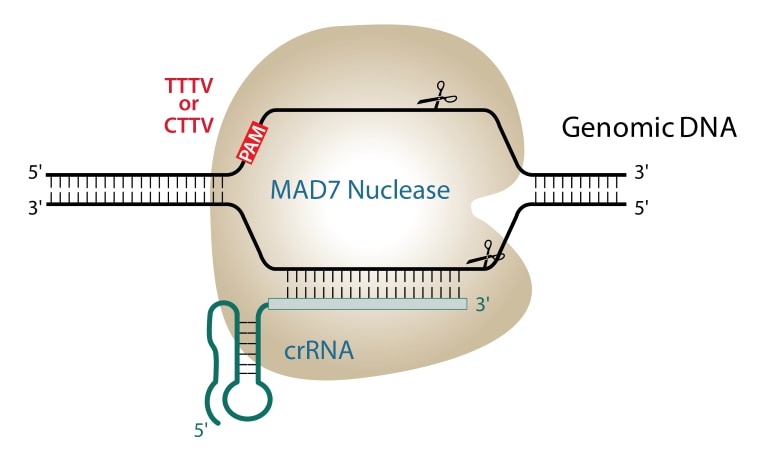
The PAM sequence is YTTV. Y indicates a C or T base, and V indicates A, C or G. The PAM sequence is located upstream of the target sequence, and the repeat sequence appended to the 5’ of the target sequence is TTAATTTCTACTCTTGTAGAT.
The DNA cleavage sites for MAD7 relative to the target site are 19 bases after the YTTV PAM site on the sense strand and 23 bases after the ecomplementary PAM site of the anti-sense strand.
When designing experiments it is always important to include controls, the recommended crRNA control sequences for use in human cell lines are below:
| Human PPIB | 5’-UUAAUUUCUACUCUUGUAGAUACCUACGAAUUGGAGAUGAAG-3’ |
| Human DNMT3B | 5’-UUAAUUUCUACUCUUGUAGAUAAAUGAACCCUGCGCUGUCAU-3’ |
These can be easily ordered through the CRISPR design tool by selecting ‘Input my own guide RNA sequence’ from the Start tab.