- Gene editing
- Gene editing reagents
- Cas9 Nuclease mRNA
Cas9 nuclease mRNA
A DNA-free option for Cas9 nuclease expression
Purified Cas9 nuclease mRNA for co-transfection with synthetic guide RNA for a completely DNA-free genome engineering system.

Cas9 nuclease in the CRISPR-Cas9 system
The CRISPR-associated enzyme Cas9 is an RNA-guided endonuclease that requires a guide RNA for genomic DNA target recognition and generation of DNA double-strand breaks (DSB).
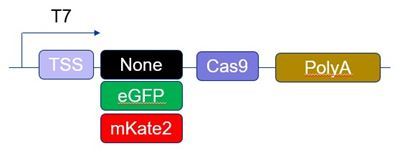
Cas9 nuclease mRNA expresses a human codon-optimized version of the S. pyogenes Cas9 nuclease gene with two nuclear localization signals (NLS). The Cas9 nuclease complexes with a guide RNA for genomic DNA target recognition and cleavage, enabling researchers to perform genome engineering experiments in a completely DNA-free manner.
Cas9 Nuclease mRNA is available in a form that co-expresses either mKate2 or EGFP for transfection optimization or enrichment using fluorescence-activated cell sorting (FACS).
Edit-R Cas9 mRNA highlights
- DNA-free systems eliminates the possibility of incorporating unwanted, exogenous DNA into the host cell's genome
- Fluorescent Cas9 mRNA enables enrichment of edited cell population (see supporting data)
- Transient expression of Cas9 nuclease to reduce off-targeting for more specific results
- No incompatibility issues between Cas9 promoter and cell line
- Easily co-transfect Edit-R synthetic guide RNA and Cas9 mRNA using DharmaFECT Duo transfection reagent, or co-electroporate into cells that are difficult-to-transfect.
Gene knockout workflow using Cas9 nuclease mRNA & Edit-R synthetic guide RNA
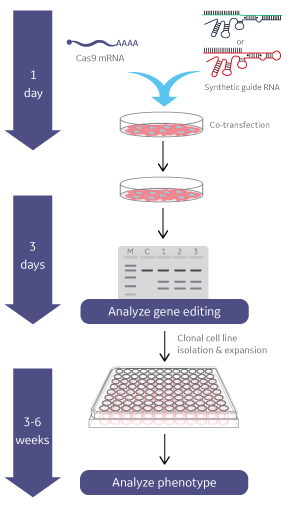
Gene editing with Cas9 Nuclease mRNA and synthetic gRNA or crRNA and tracrRNA is performed by co-transfecting all components with DharmaFECT Duo Transfection Reagent (or other DharmaFECT transfection reagent suitable to your specific cells of interest). One may then observe phenotypes directly. A DNA mismatch detection assay can be used to estimate gene editing efficiency prior to clonal cell line generation and characterization.
Fluorescent Cas9 nuclease mRNA data
Fluorescent Cas9 nuclease mRNA maintains editing efficiency
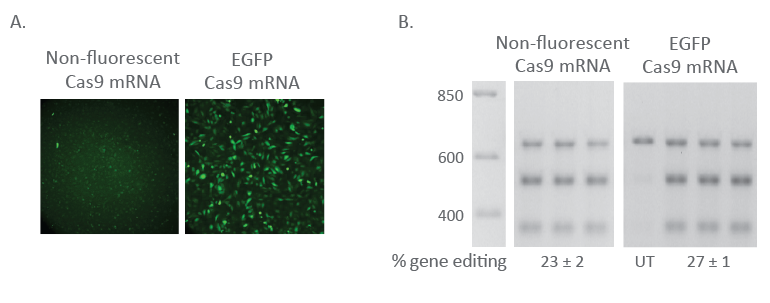
Fluorescent Cas9 mRNA U2OS cells were plated at 10,000 cells/well in a black 96-well plate 24 hours before transfection. At the time of transfection, EGFP Cas9 nuclease mRNA (200 ng, Cat #CAS11860) or non-fluorescent Cas9 Nuclease mRNA (200 ng, Cat #CAS11195) was co-transfected with Edit-R PPIB Synthetic crRNA Control (25 nM, Cat #U-007000-01-05) and tracrRNA (25 nM, Cat# U-002005-05) using DharmaFECT™ Duo transfection reagent (0.3 µL/well). After 24 hours, cells were imaged for EGFP fluorescence using the Nikon Eclipse Ti-S/L100 inverted microscope (A). At 72 hours after transfection, cells were harvested and gene editing was measured through a DNA mismatch detection assay (B).
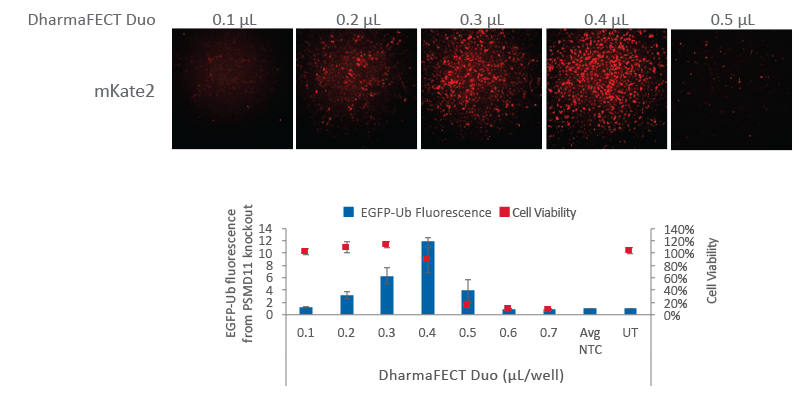
Cas9 nuclease mRNA with fluorescent reporter indicates optimal transfection conditions
U2OS cells expressing a Ubiquitin-tagged EGFP were plated at 10,000 cells/well in a black 96-well plate and co-transfected with Cas9 mRNA (200 ng, Cat #CAS11859) and synthetic tracrRNA (25 nM, Cat# U-002005-05) targeting PSMD11 (25 nM, Cat #CM-085161-03), a known proteasome component, or a Non-targeting control (NTC, Cat #U-007501-xx). Increasing amounts of DharmaFECT Duo Transfection Reagent were tested per well (0.1 µL to 0.7 µL). After 24 hours, cells were imaged for mKate2 fluorescence using the In Cell Analyzer 2200. At 72 hours after transfection, cells were imaged for EGFP fluorescence resultant from proteasome knockout. Based on mKate2 fluorescence, optimal transfection conditions occur at 0.3-0.4 µL of DharmaFECT Duo per well which corresponds to an increased phenotypic effect. Above 0.4 µL of DharmaFECT Duo per well, significant cell death is observed (graph, red boxes). UT = untransfected sample.
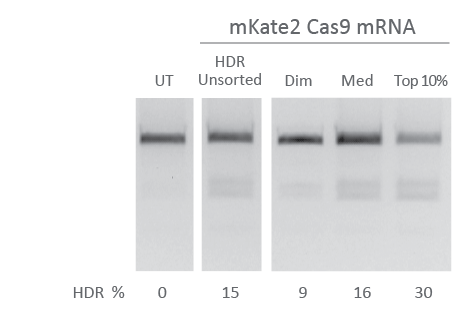
Fluorescent Cas9 mRNA enables enrichment of HDR edited cells
U2OS cells were transfected with Edit-R mKate2-Cas9 mRNA (200 ng, Cat #CAS11859), EMX1-targeting crRNA (25 nM, Custom synthesis), tracrRNA (25 nM, Cat #U-002005) and a single-stranded donor oligo (5 nM, designed with the Edit-R HDR Donor Designer) to insert a NheI restriction site and a FLAG tag sequence at the C-terminus of EMX1. Transfections were done using DharmaFECT Duo transfection reagent (0.3 µL/well) 24 hours after plating 10,000 U2OS cells per well of a 96-well plate. Cells were then collected 24 hours after transfection and sorted into mKate2 fluorescent subpopulations (either Dim mKate2 fluorescence, Medium, or Top 10% fluorescence). Subpopulations were expanded for an additional 48 hours and analyzed for NheI insertion through Restriction Fragment Length Polymorphism (RFLP) by treating EMX1 PCR amplicons with NheI restriction enzyme.
Cas9 nuclease mRNA data
Cas9 nuclease mRNA gene editing in three cell lines
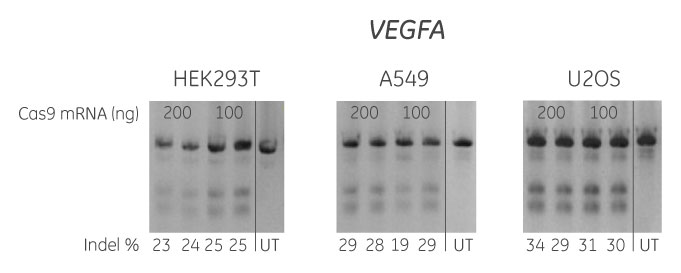
HEK293T, A549 and U2OS cells were plated at 20,000 cells/well in 96-well plates and co-transfected using DharmaFECT Duo Transfection Reagent with Cas9 mRNA (200 or 100 ng) and synthetic crRNA:tracrRNA (50 nM) targeting VEGFA. Cells were harvested 72 hours post-transfection and the relative frequency of gene editing was calculated based on a DNA mismatch detection assay with T7 Endonuclease I. The synthetic crRNA was designed using the Dharmacon CRISPR Design Tool. UT = untransfected sample.
Cas9 nuclease mRNA dose curve for efficient gene editing
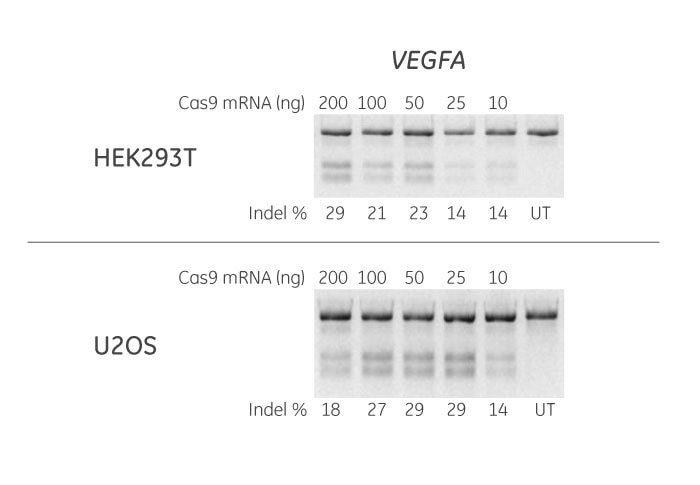
HEK293T and U2OS cells were plated at 20,000 cells/well in 96-well plates and co-transfected using DharmaFECT Duo Transfection Reagent with Cas9 mRNA (200, 100, 50, 25 or 10 ng) and synthetic crRNA:tracrRNA (50 nM) targeting VEGFA. Cells were harvested 72 hours post-transfection and the relative frequency of gene editing was calculated based on a DNA mismatch detection assay with T7 Endonuclease I. It is recommended to optimize the amount of Cas9 mRNA (50-200 ng/well in 96-well format) in your cells of interest and for your application. The synthetic crRNA targeting VEGFA was designed using the Dharmacon CRISPR Design Tool. UT = untransfected sample.
Efficiency of gene editing with 2x MS modified or unmodified crRNA:tracrRNA
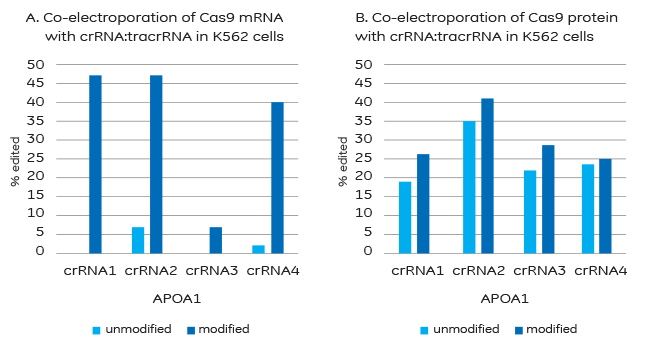
A. Modifications to block degradation by nucleases are required for successful co-electroporation of Cas9 mRNA and crRNA:tracrRNA. B. Co-electroporation of Cas9 protein and crRNA:tracrRNA does not require stabilizing modifications, but results may be improved with their addition.
Application notes
-
Fluorescent Cas9 mRNA for enrichment of CRISPR-mediated knockout and knock-in using synthetic guide RNA - Application Note
-
Functional gene knockout in human iPS cells using Edit-R CRISPR reagents - Application Note
-
Microinjection of zebrafish embryos using Dharmacon Edit-R Cas9 Nuclease mRNA, synthetic crRNA, and tracrRNA for genome engineering - Application Note
-
Using the CRISPR-Cas9 system with paired Dharmacon Edit-R synthetic crRNAs for functional knockout of microRNA hsa-miR-221 - Application Note
Posters
Protocols
-
Cas9 Nuclease mRNA Electroporation - Protocol
-
Fluorescent Cas9 Nuclease mRNA for enrichment of transfected cells - Protocol
-
T7E1, TIDE, and NGS analysis protocol for Dharmacon™ Edit-R™ gene editing experiments
-
Transfection of ssDNA donor oligonucleotides for HDR-mediated gene alterations using the Dharmacon Edit-R system - Protocol