- CRISPR activation reagents
- CRISPRa synthetic crRNA non-targeting controls
CRISPRmod CRISPRa synthetic crRNA non-targeting controls

CRISPRa crRNA non-targeting controls are designed and recommended for use as negative controls for transcriptional activation experiments using CRISPRa crRNA individuals and pools. These non-targeting controls will hybridize with tracrRNA and engage the dCas9-VPR complex, but will not target any PAM-adjacent sites in the human or mouse genome. Any observed alteration in gene expression levels or viability in cells treated with these controls can be used as a baseline response of the cells to the dCas9-VPR complex for comparison to those treated with target-specific CRISPRa crRNAs and pools.
Highlights
- Proprietary alignment tools used to verify at least three mismatches or gaps to any potential target in human or mouse genomes.
- Choose an individual or pooled crRNA control to match your experimental crRNA reagent.
Please note that tracrRNA is required for use with CRISPRa synthetic crRNA reagents.
| crRNA nmol | tracrRNA nmol | 96-well plate 100 µL volume | 24-well plate 500 µL volume | 12-well plate 1000 µL volume | 6-well plate 2500 µL volume |
|---|---|---|---|---|---|
| 2 | 2 | 800 | 160 | 80 | 32 |
| 5 | 5 | 2000 | 400 | 200 | 80 |
| 10 | 10 | 4000 | 800 | 400 | 160 |
| 20 | 20 | 8000 | 1600 | 800 | 300 |
CRISPRa workflow diagram with stable dCas9-VPR expression
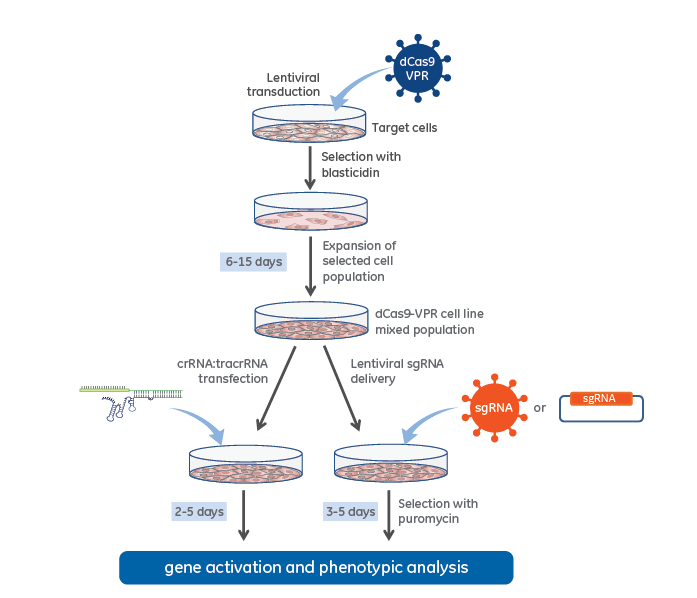
CRISPR activation workflow with lentiviral dCas9-VPR and synthetic crRNA:tracrRNA (left side) or Lentiviral expressed sgRNA (right side).
Pooling of CRISPRa synthetic crRNAs enhances transcriptional activation
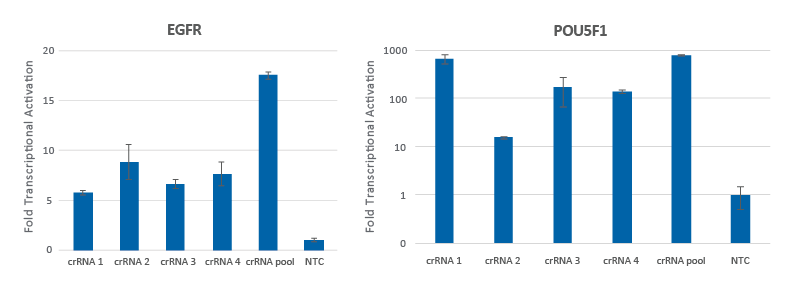
Individual CRISPRa crRNAs achieve robust target gene activation on their own, but when pooled together in a single reagent, enhanced activation levels can be achieved. U2OS cells stably expressing integrated CRISPRa dCas9-VPR were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA targeting EGFR or POU5F1. Four CRISPRa crRNAs were used either individually or pooled (to a total concentration of 25 nM). Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative fold transcriptional activation for each gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).
Application notes
Safety data sheets
Related Products
Chemically synthesized trans-activating CRISPR RNA required for use with synthetic crRNA for fast and easy gene editing
Purified lentiviral particles or plasmid DNA for generation of stable dCas9-VPR nuclease-expressing cell populations
Validated synthetic crRNA pool or individual for experimental optimization of transcription activation experiments.
