- Dharmacon Screening libraries
- Human ON-TARGETplus siRNA Library - Ubiquitin Enzymes
Human ON-TARGETplus siRNA Library - Ubiquitin Enzymes
siRNA designed and modified for greater specificity

The Human ON-TARGETplus Ubiquitin Enzymes Library combines gene targets from four of our ubiqutin-related libraries into one comprehensive collection for investigation of regulators of the ubiqutination pathway:
- Deubiquitinating Enzymes
- Ubiquitin Conjugation Subset 1 - Cullins, E1, E2, HECT E3 Ligases
- Ubiquitin Conjugation Subset 2 - F-box, SOCS box E3 Ligases
- Ubiqutin Conjugation Subset 3 - RING finger and RING finger-like E3 Ligases
ON-TARGETplus siRNA reagents are globally recognized and trusted for highly effective performance in RNAi screening applications. Our expertise in siRNA design, experimental optimization, and siRNA screening strategies provide end-to-end support of your screening efforts.
Highlights
- Patented dual-strand ON-TARGETplus modification pattern on all siRNAs to reduce off-targets
- Sense strand is modified to prevent interaction with RISC and favor antisense strand uptake
- Antisense strand seed region is modified to destabilize off-target activity and enhance target specificity
- Available as SMARTpool siRNA reagents or a Set of 4 siRNAs in 384-well plates
- Guaranteed target gene knockdown (see Specifications tab)
Our siRNA knockdown guarantee
siGENOME and ON-TARGETplus siRNA reagents (SMARTpool and three of four individual siRNAs) are guaranteed to silence target gene expression by at least 75% at the mRNA level when demonstrated to have been used under optimal delivery conditions (confirmed using validated positive control and measured at the mRNA level 24 to 48 hours after transfection using 100 nM siRNA).
Note: Most siGENOME and ON-TARGETplus siRNA products are highly functional at 5 to 25 nM working concentration.
Gene Targets
For a complete list of target genes in this siRNA Library, please contact Scientific Support or your local Sales Representative.
Our siRNA knockdown guarantee
siGENOME and ON-TARGETplus siRNA reagents (SMARTpool and three of four individual siRNAs) are guaranteed to silence target gene expression by at least 75% at the mRNA level when demonstrated to have been used under optimal delivery conditions (confirmed using validated positive control and measured at the mRNA level 24 to 48 hours after transfection using 100 nM siRNA).
Note: Most siGENOME and ON-TARGETplus siRNA products are highly functional at 5 to 25 nM working concentration.
False phenotypes due to off-targets are alleviated by ON-TARGETplus SMARTpool reagents while target gene knockdown is maintained
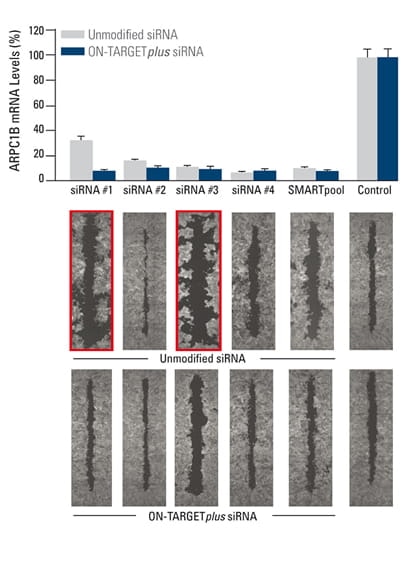
The effect of silencing ARPC1B on cell migration was studied in a breast cancer cell line. A monolayer of cells was uniformly scraped and the rate of cell migration to close the scrape (wound healing) was evaluated. Both unmodified and ON-TARGETplus siRNA reagents induced potent target knockdown. Inconsistent phenotypes due to off-target effects (red outline), were observed for cells transfected with unmodified individual siRNAs. The unmodified SMARTpool improved the false phenotype considerably, while the ON-TARGETplus SMARTpool significantly reduced off-target effects to produce a consistent phenotype. In collaboration with Kaylene Simpson, Laura Selfors, and Joan Brugge, Harvard Medical School.
ON-TARGETplus modifications reduce the overall number of off-targets and pooling reduces them even further
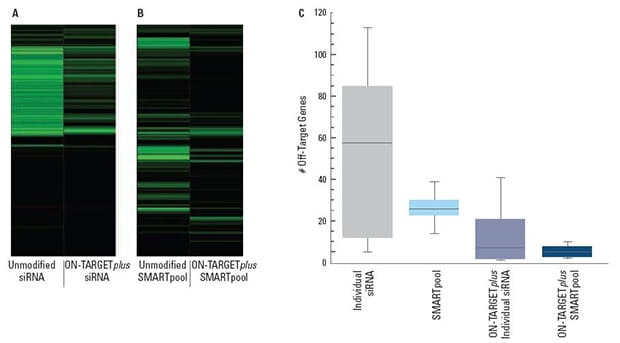
(A) and (B) are representative examples of off-target signatures with and without application of ON-TARGETplus modifications to (A) a single siRNA and (B) a SMARTpool reagent. Green bars indicate genes with 2-fold or more reduction of expression when treated with the indicated siRNA reagent. The ON-TARGETplus modifications reduced the off-targets when compared to unmodified siRNA. Pooling of siRNA and the ON-TARGETplus modification pattern independently, and in combination, provide significant reduction in off-target gene silencing. Panel (C) represents quantitation of off-targets (down-regulated by 2-fold or more) induced by the indicated siRNA reagents targeting 10 different genes (4 siRNAs per gene or a single SMARTpool reagent). Off-targets were quantified using microarray analysis (Agilent) then compiled. Each shaded box represents the middle 50% of the data set. Horizontal line in box: Median value of the data set. Vertical bars: minimum and maximum data values.
- B. D. Parsons et al., A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS One. 4(12), e8471 (29 December 2009).
- M. Jiang et al., Tales from an academic RNAi screening facility; FAQs. Brief Funct Genomics. 10(4), 227-237 (Epub 28 April 2011, July 2011). [doi: 10.1093/bfgp/elr016]
- A. Genovesio et al., Visual genome-wide RNAi screening to identify human host factors required for Trypanosoma cruzi infection. PLoS One. 6(5), e19733 (Epub 20 May 2011).
- J. A. Smith et al., Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U S A. 107(8), 3752-3757. (23 February 2010).
- G. Hu et al., A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23(7), 837-848 (1 April 2009).
- R. D. Paulsen et al., A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 35(2), 228-239 (31 July 2009).
- M. Tsui et al., An intermittent live cell imaging screen for siRNA enhancers and suppressors of a kinesin-5 inhibitor. PLoS One. 4(10), e7339 (5 October 2009).
- A. L. Brass et al., Identification of host proteins required for HIV infection through a functional genomic screen. Science. 319(5865), 921-926. (15 February 2008).
- C. Swanton et al., Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 11(6), 498-512 (11 June 2007).
- A. W. Whitehurst et al., Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 446(7137), 815-819 (12 April 2007).
