- Dharmacon Screening libraries
- Mouse siGENOME siRNA Library - Ubiquitin Conjugation Subset 1
Mouse siGENOME siRNA Library - Ubiquitin Conjugation Subset 1
Trusted for superior silencing since 2002

The Mouse siGENOME Ubiquitin Conjugation Subset 1 siRNA library targets Cullin, E1, E2, and HECT E3 ligases. Assessment of effects of E1 and E2 enzymes will provide a reasonable idea as to whether components of the ubiquitin system are involved.
siGENOME siRNA reagents are globally recognized and trusted for highly effective performance in RNAi screening applications. Our expertise in siRNA design, experimental optimization, and siRNA screening strategies provide end-to-end support of your screening efforts.
HECT domain E3 ligases, included in this set, differ from the large majority of known E3s (RING finger and RING finger-like) in functioning as catalytic intermediates in ubiquitin conjugation. Members of this family are implicated in the ubiquitylation of many different proteins. Some of these events lead to proteasomal degradation while others are implicated in non-proteasomal ubiquitylation, including trafficking within the endosomal pathway.
Highlights
- Guaranteed target knockdown (see Specifications tab)
- Antisense strand loading into RISC ensured by thermodynamic analysis and selective application of a sense strand-blocking modification (ON-TARGET)
- Available as SMARTpool or Set of 4 siRNA reagents arrayed in 96-well plates
Gene Targets
For a complete list of target genes in this siRNA Library, please contact Scientific Support or your local Sales Representative.
For a thorough investigation of the ubiquitin pathway, you may also consider these additional libraries:
Our siRNA knockdown guarantee
siGENOME and ON-TARGETplus siRNA reagents (SMARTpool and three of four individual siRNAs) are guaranteed to silence target gene expression by at least 75% at the mRNA level when demonstrated to have been used under optimal delivery conditions (confirmed using validated positive control and measured at the mRNA level 24 to 48 hours after transfection using 100 nM siRNA).
Note: Most siGENOME and ON-TARGETplus siRNA products are highly functional at 5 to 25 nM working concentration.
Screening with siGENOME SMARTpool siRNA reagents provides effective target silencing
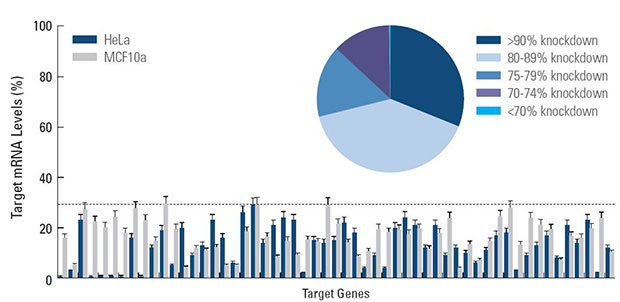
Effective silencing achieved for 440 kinase genes when screened with SMARTpool siRNA reagents in two cell lines. Under screening conditions these overall results provide validation of siRNA design and SMARTpool technology for effective target knockdown. HeLa and MCF10a cells were transfected with 50 nM of each siRNA using DharmaFECT 1 transfection reagent. Target mRNA levels were determined by branched DNA assay (Panomics Quantigene® Reagent System). The pie chart represents the summary of silencing achieved for the entire data set (880 total data points). The bar graph is a representative sample of genes assayed.
Unnecessary sense-strand inactivation can increase off-target activity
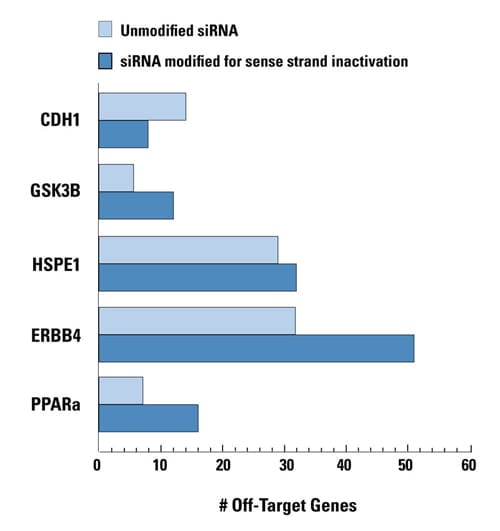
Unmodified and sense strand-inactivated siRNAs were used to target five genes. In four cases, off-targets were increased due to enhanced RISC loading of the antisense strand when the sense strand was modified. The unmodified siRNAs have natural guide-strand loading characteristics. All siRNAs had comparable silencing potency. Data shown represents genes down-regulated by twofold or more. HEK293 cells were transfected with 100 nM siRNA using 0.2 μL of DharmaFECT 1. Data was analyzed at 24 hours by genomewide microarray analysis (Agilent).
Why not modify ALL siGENOME siRNAs to ensure proper strand loading? It has been demonstrated by Dharmacon scientists1 and others2 that forcing antisense (guide) strand entry into RISC may actually INCREASE off-targets due to increased loading of the guide strand and resulting off-target activity by its seed region. siGENOME siRNAs are designed with thermodynamic properties to naturally facilitate guide strand entry to RISC, which has been demonstrated to correlate with functionality1. However, in cases where a high-scoring siGENOME siRNA does not possess ideal strand-loading characteristics, a sense (passenger) strand-inhibiting chemical modification (ON-TARGET) is utilized to promote guide strand entry. Approximately 20% of siGENOME siRNAs carry the ON-TARGET modification.
- B. D. Parsons et al., A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS One. 4(12), e8471 (29 December 2009).
- M. Jiang et al., Tales from an academic RNAi screening facility; FAQs. Brief Funct Genomics. 10(4), 227-237 (Epub 28 April 2011, July 2011). [doi: 10.1093/bfgp/elr016]
- J. Borawski et al., Optimization procedure for small interfering RNA transfection in a 384-well format. J Biomol Screen. 12(4), 546-559 (Epub 13 April 2007, June 2007).
- J. D. Zhang et al., Time-resolved human kinome RNAi screen identifies a network regulating mitotic-events as early regulators of cell proliferation. PLoS One. 6(7), e22176 (Epub 13 July 2011).
- A. Genovesio et al., Visual genome-wide RNAi screening to identify human host factors required for Trypanosoma cruzi infection. PLoS One. 6(5), e19733 (Epub 20 May 2011).
- J. A. Smith et al., Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U S A. 107(8), 3752-3757. (Epub 2 February 2010, 23 February 2010).
- G. Hu et al., A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23(7), 837-848 (1 April 2009).
- R. D. Paulsen et al., A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 35(2), 228-239 (31 July 2009).
- M. Tsui et al., An intermittent live cell imaging screen for siRNA enhancers and suppressors of a kinesin-5 inhibitor. PLoS One. 4(10), e7339 (5 October 2009).
- A. L. Brass et al., Identification of host proteins required for HIV infection through a functional genomic screen. Science. 319(5865), 921-926. (Epub 10 January 2008, 15 February 2008).
- C. Swanton et al., Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 11(6), 498-512 (11 June 2007).
- A. W. Whitehurst et al., Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 446(7137), 815-819 (12 April 2007).
Product inserts
Related Products
Concentrated buffer solution recommended for resuspension and long-term storage of any short, double-strand, or single-strand synthetic RNA molecule. Dilute with RNase-free water prior to use.
Catalog ID:B-002000-UB-100
Unit Size:100 mL
$99.00
Molecular grade water for dilution of 5x siRNA Buffer or resuspension of RNA. RNase-free to prevent degradation of RNA reagents and oligonucleotides.
Catalog ID:B-003000-WB-100
Unit Size:100 mL
$32.00
An aliquot of each of the four DharmaFECT formulations for siRNA/microRNA transfection optimization studies
Catalog ID:T-2005-01
Unit Size:0.2 mL
