What is shRNA and how do you use it?
Short hairpin RNA (shRNA) sequences are usually encoded in a DNA vector that can be introduced into cells via plasmid transfection or viral transduction. shRNA molecules can be divided into two main categories based on their designs: simple stem-loop and microRNA-adapted shRNA. A simple stem-loop shRNA is often transcribed under the control of an RNA Polymerase III (Pol III) promoter1, 2. The 50-70 nucleotide transcript forms a stem-loop structure consisting of a 19 to 29 bp region of double-strand RNA (the stem) bridged by a region of predominantly single-strand RNA (the loop) and a dinucleotide 3' overhang 3-5. The simple stem-loop shRNA is transcribed in the nucleus and enters the RNAi pathway similar to a pre-microRNA. The longer (> 250 nucleotide) microRNA-adapted shRNA is a design that more closely resembles native pri-microRNA molecules, and consists of a shRNA stem structure which may include microRNA-like mismatches, bridged by a loop and flanked by 5' and 3' endogenous microRNA sequences6. The microRNA-adapted shRNA, like the simple stem-loop hairpin, is also transcribed in the nucleus but is thought to enter the RNAi pathway earlier similar to an endogenous pri-microRNA.
shRNA technologies are DNA-based, which provides flexibility in design of the vector. Most vector-based shRNA systems contain a selectable marker to allow for the elimination of cells that have not been successfully transfected or transduced, and maintenance of cells with sustained gene knockdown. The shRNA expression cassettes can also be incorporated into viral vector systems, including retrovirus, adeno-associated virus, adenovirus and lentivirus, which permit stable integration into and expression from the host genome. These viral strategies permit shRNA delivery to cell lines that are refractory to transfection. Fluorescent markers (such as a Green or Red Fluorescent Protein [GFP or RFP]) can also be included for tracking cells expressing shRNAs. The performance of shRNA is influenced by many factors including the efficiency of transduction or transfection, the promoter driving expression of the shRNA and epigenetic modifications (which can lead to silencing of shRNA expression). Further, the influence that each of these factors have on vector performance can differ depending on the cell line or cell type7-9. SMARTchoice promoter and reporter options are available to help researchers select optimal promoters for shRNA expression and gene silencing. Finally, when these shRNA expression cassettes are coupled with inducible promoters, as with SMARTvector Inducible Lentiviral shRNA or the TRIPZ vector system, researchers can design studies to temporally and spatially modulate gene expression10-12.
How is shRNA delivered to a cell?
There are two options for DNA-based shRNA delivery to cells. For plasmids, typical transfection methods such as use of lipid transfection reagents or electroporation can be employed. Lentiviral particles are an excellent choice for difficult to transfect cells, and in situations where high efficiency is needed or multiple constructs per cell need to be delivered. Most modern vectors have selectable markers that allow the selective killing of cells in the culture that were not successfully transfected or transduced, so that a pure culture can be developed.
What affects shRNA function and specificity?
Optimal gene knockdown is a requirement for successful RNAi using shRNA systems. Rational design of shRNA sequences has largely been based on algorithms developed with siRNA. While some siRNA design rules apply to shRNA, more refined shRNA design algorithms will likely improve target gene silencing for shRNAs in the future13. In order to predict functional shRNA sequences, Dharmacon's SMARTvector™ shRNA algorithm selects target sequences on the basis of numerous criteria, including position-dependent nucleotide preferences, secondary structure and thermodynamic stability profiles specific to the SMARTvector microRNA-based scaffold. Additionally, the SMARTvector algorithm includes several criteria for increasing specificity.
As with siRNAs, bioinformatic approaches can be applied to create target-specific shRNAs while minimizing the potential for off-target effects. High concentrations of silencing intermediates are known to contribute to off-target events, but the level of the intermediates are difficult to control when shRNAs are exogenously expressed. Several publications have documented evidence that, under specific conditions, high levels of simple stem-loop shRNA expression may saturate the endogenous RNAi pathway, and lead to unintended phenotypes14. Other studies have suggested that the use of a microRNA-adapted shRNA may reduce cellular toxicity for in vivo RNAi experiments6,15 due to these scaffolds being more efficiently processed by both Drosha-DGCR8 and Dicer 16,17.
shRNA design
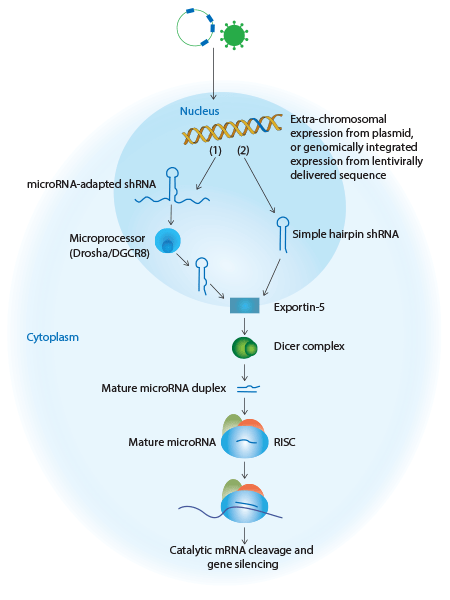
shRNA approaches include the introduction of genetically engineered viral vectors or plasmid-based vectors expressing silencing sequences embedded in an endogenous microRNA scaffold (1) or simple stem-loop shRNA (2). Expressed sequences (1 and 2, shown in blue) enter the endogenous pathway at an early stage and are efficiently processed into potent silencing molecules using the endogenous microRNA mechanism. All of these approaches lead to target mRNA cleavage (shown in purple) and gene silencing.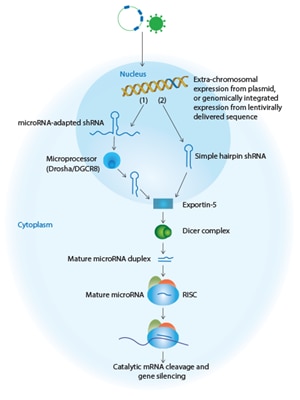
RNA interference and manipulation
RNA interference and manipulation
Simple stem-loop shRNA
Basic shRNAs are modeled on precursor microRNA (pre-miRNA), and are cloned into viral vectors where they are transcribed under the control of RNA Polymerase III (Pol III) promoters. shRNAs are produced as single-strand molecules of 50–70 nucleotides in length, and form stem loop structures consisting of a 19-29 base-pair region of double-strand RNA (the stem) bridged by a region of single-strand RNA (the loop) and a short 3’ overhang. Once transcribed, shRNAs exit the nucleus, are cleaved at the loop by the nuclease Dicer in the cytoplasm, and enter the RISC to direct cleavage and subsequent degradation of complementary mRNA.
microRNA adapted shRNA
A microRNA-adapted shRNA consists of a shRNA stem structure with microRNA-like mismatches surrounded by the loop and flanking sequence of an endogenous microRNA. microRNA-adapted shRNAs are transcribed from RNA Polymerase ll (Pol ll) promoters, cleaved by the endogenous RNase III Drosha enzyme in the nucleus, and then exported to the cytoplasm where they are processed by Dicer and loaded into the RISC complex. Studies have suggested that the use of a microRNA scaffold, which is processed by both Drosha and Dicer, may promote more efficient processing and reduce toxicity for in vivo RNAi.
shRNA applications
shRNA offers the possibility of prolonged gene silencing. Transduction of viral-based shRNA allows access to cells, such as primary and neuronal cells, that are difficult to transfect by traditional cationic lipid-based strategies. Viral-based shRNAs have also been used to evaluate gene function at whole genome scales using pools of silencing constructs. Pooled or arrayed screens are now widely used to perform high-throughput screens to identify genes required in a variety of processes such as cancer cell survival and proliferation 18-20, tumor suppressor pathway components21, modulators of the mammalian circadian clock22, suppressors of epithelial mesenchymal transition23, host mediators of HIV-1 replication24,25, and regulators of cell migration26. The power of pooled RNAi screening has also been extended to screening for gene function in animal models to investigate in vivo biolog27, 28.
References
- Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281-297.
- Kim, V.N. (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews, Molecular Cell Biology 6(5):376-385.
- Brummelkamp, T.R. et al. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296(5567):550-553.
- Paddison, P.J. et al. (2002) Stable suppression of gene expression by RNAi in mammalian cells. PNAS 99(3):1443-1448.
- Paul, C.P. et al. (2002) Effective expression of small interfering RNA in human cells. Nature Biotechnology 20(5):505-508.
- Silva, J.M. et al. (2005) Second-generation shRNA libraries covering the mouse and human genomes. Nature Genetics 37(11):1281-1288.
- Hong, S. et al. (2007) Functional analysis of various promoters in lentiviral vectors at different stages of in vitro differentiation of mouse embryonic stem cells. Molecular Therapy 15(9):1630-1639.
- Liu, Z. et al. (1997) A systematic comparison of relative promoter/enhancer activities in mammalian cell lines. Analytical Biochem. 246(1):150-152.
- Ramezani, A. et al. (2000) Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Molecular Therapy 2(5):458-469.
- Ying, M. et al. (2011) Kruppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch2 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells 29(1):20-31.
- Peng, J. et al. (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139(7):1290-1302.
- Du, W. et al. (2010) Cytoplasmic FANCA-FANCC complex interacts and stabilizes the cytoplasm-dislocalized leukemic Nucleophosmin Protein (NPMc). Journal of Biological Chemistry 285(48):37436-37444.
- Fellmann, C. et al. (2011) Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Molecular Cell 41(6):733-746.
- Grimm, D. et al. (2006) Fatality in mice due to oversaturation of cellular microRNA/ short hairpin RNA pathways. Nature 441:537-541.
- McBride, J.L. et al. (2008) Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. PNAS 105(15):5868-5873.
- Siolas, D. et al. (2005) Synthetic shRNAs as potent RNAi triggers. Nature Biotechnology 23(2):227-231.
- Gregory, R.I. et al. (2005) Human RISC couples microRNA biogenesis and post-transcriptional gene silencing. Cell 123(4):631-640.
- Luo, B. et al. (2008) Highly parallel identification of essential genes in cancer cells. PNAS 105(51):20380-20385.
- Silva, J.M. et al. (2008) Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319(5863): 617-620.
- Schlabach, M.R. et al. (2008) Cancer proliferation gene discovery through functional genomics. Science 319(5863):620-624.
- Mullenders, J. et al. (2009) Candidate biomarkers of response to an experimental cancer drug identified through a large-scale RNA interference genetic screen. Clinical Cancer Research 15(18):5811-5819.
- Maier, B. et al. (2009) A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes and Development 23:708-718.
- Gumireddy, K. et al. (2009) KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nature Cell Biology 11(11):1297-1304.
- Rato, S. et al. (2010) Novel HIV-1 knockdown targets identified by an enriched kinases/phosphatases shRNA library using a long-term iterative screen in Jurkat T-cells. PLoS One 5(2):e9276.
- Yeung, M.L. et al. (2009) A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. Journal of Biological Chemistry 284(29):19463-19473.
- Smolen, G.A. et al. (2010) A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes and Development 24(23):2654- 2665.
- Zender, L. et al. (2008) An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135(5):852-864.
- Montgomery, R.L. et al. (2011) Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 124(14):1537-1547.
Which shRNA tool is right for your needs?
shRNA Selection Guide
We offer a selection of shRNA reagents and libraries. This quick selection guide will help to determine the best option for your particular needs.
Order Products
shRNA Product Overview
Lentiviral vector-based reagents for RNA interference
SMARTvector Lentiviral shRNA
Make smart, informed choices for successful gene silencing in your cells of interest
SMARTvector Inducible shRNA
The most advanced and flexible single-vector inducible shRNA available for tightly controlled gene silencing
Pooled Lentiviral Screening Libraries
Optimized construction of high-quality pools, complete analysis tools, and validated protocols are essential to a successful outcome when screening with pooled lentiviral libraries
Helpful Resources
SMARTvector Lentiviral shRNA - Tech Note
Technical note describing the SMARTchoice shRNA experimental workflow in suspension cells
SMARTvector Lentiviral shRNA - Technical Manual
The platform is an innovative system ideally suited for RNAi-mediated studies
GIPZ Lentiviral shRNA - Technical Manual
Experimental protocols for gene knockdown using GIPZ lentiviral shRNA
