- CRISPR activation reagents
- CRISPRa synthetic crRNA
CRISPRmod CRISPRa synthetic crRNA
Predesigned synthetic guide RNA for over-expression of human and mouse genes using CRISPR activation. Just search for your gene! Available as pooled or individual reagents.

CRISPRa synthetic crRNA
1Start Here
2Choose
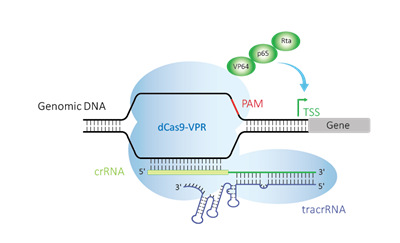
Endogenous gene activation with an easy-to-use synthetic reagent
The CRISPR activation (CRISPRa) system is a unique adaptation of the classical CRISPR-Cas9 gene editing system which utilizes a catalytically deactivated Cas9 (dCas9) that is fused to one or more transcriptional activators. When paired with a well-designed guide RNA that targets a gene near a promoter region, or trancriptional start site (TSS), it promotes transcriptional activation.
Review our CRISPRa application page to get an overview of the technology!
Highlights of CRISPRa crRNA reagents
- Available as individual reagents or a pool of four crRNAs to provide highly effective transcriptional activation (See Supporting Data tab).
- Designs from a published algorithm by Horlbeck, et. al. that demonstrates strong levels of gene activation from optimized designs (See References tab).
- Chemically modified crRNAs provide additional stability against nuclease degradation and improve overall performance.
- For genes with alternative characterized start sites, distinct guide RNA designs are available (labeled P2).
Required components for CRISPRa gene activation using synthetic crRNA
- A lentiviral expression plasmid, lentiviral particles or mRNA for dCas9-VPR; a mammalian codon-optimized S. pyogenes deactivated Cas9 fused to VPR activation domains (also compatible with SunTag technology).
- A chemically synthesized trans-activating CRISPR RNA (tracrRNA).
- A predesigned CRISPRa crRNA targeting the gene of interest.
How much CRISPRa crRNA & tracrRNA do I need?
This table provides the approximate number of experiments that can be carried out for lipid transfection methods at the recommended crRNA:tracrRNA working concentration (25 nM:25nM) in various plate/well formats using either a pool or individual crRNA. Calculations do not account for pipetting errors.
| crRNA nmol | tracrRNA nmol | 96-well plate 100 µL volume | 24-well plate 500 µL volume | 12-well plate 1000 µL volume | 6-well plate 2500 µL volume |
|---|---|---|---|---|---|
| 2 | 2 | 800 | 160 | 80 | 32 |
| 5 | 5 | 2000 | 400 | 200 | 80 |
| 10 | 10 | 4000 | 800 | 400 | 160 |
| 20 | 20 | 8000 | 1600 | 800 | 300 |
CRISPRa workflow diagram with stable dCas9-VPR expression
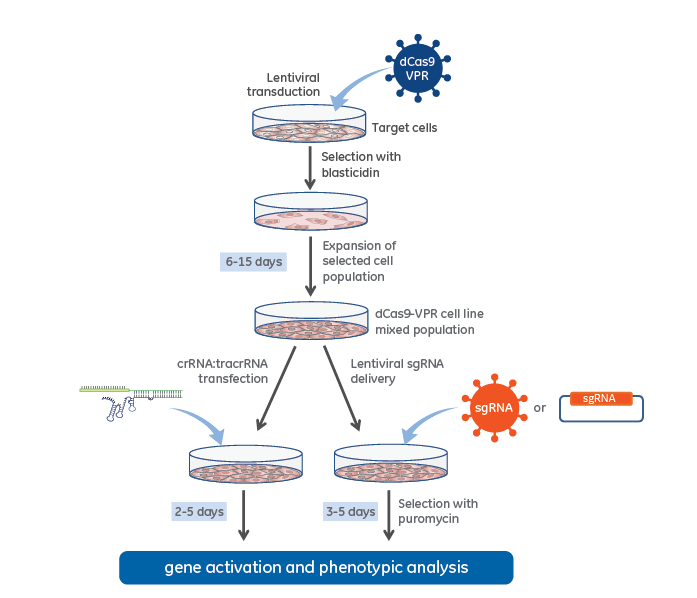
CRISPR activation workflow with lentiviral dCas9-VPR and synthetic crRNA:tracrRNA (left side) or Lentiviral expressed sgRNA (right side).
CRISPRa controls
CRISPRa synthetic crRNA positive controls
- CRISPRa crRNAs targeting well-characterized genes to determine the effectiveness of your experimental conditions for maximum activation.
CRISPRa synthetic crRNA non-targeting controls
- Non-targeting controls to evaluate baseline cellular responses to CRISPRa components in the absence of gene target-specific crRNA.
CRISPRa related products
Synthetic tracrRNA
- Trans-activating CRISPR RNA (tracrRNA) is a synthetic, HPLC-purified, long RNA required for use with crRNA to form the complex that programs dCas-VPR. It is modified for nuclease resistance.
CRISPRa dCas9-VPR
- Lentiviral particles, purified plasmid or mRNA that express nuclease-deactivated Cas9 fused to transcriptional activators. When complexed with a guide RNA will trigger an endogenous gene’s expression.
Efficient gene activation with CRISPRa synthetic crRNA in multiple dCas9-VPR stable cell lines
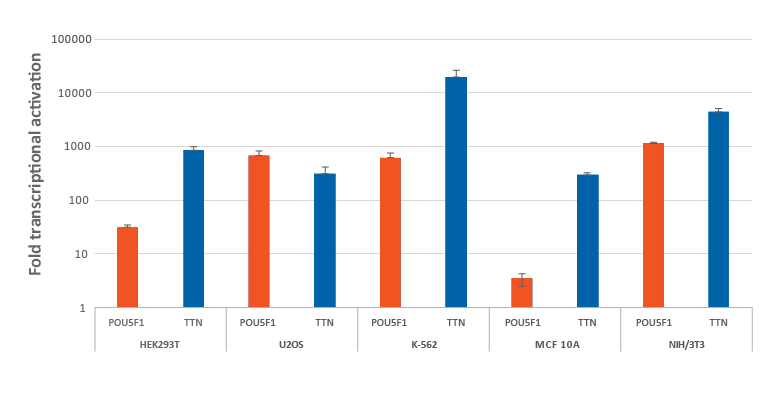
HEK293T, U2OS, MCF 10A, NIH/3T3 stably expressing integrated CRISPRa dCas9-VPR were plated at 10,000 cells/well and transfected using DharmaFECT Transfection Reagents with CRISPRa synthetic crRNA:tracrRNA (25 nM) targeting POU5F1 and TTN. K562 cells were electroporated with CRISPRa synthetic crRNA:tracrRNA (400 nM) targeting POU5F1 and TTN. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative fold transcriptional activation for each gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
Pooling of CRISPRa synthetic crRNAs enhances transcriptional activation
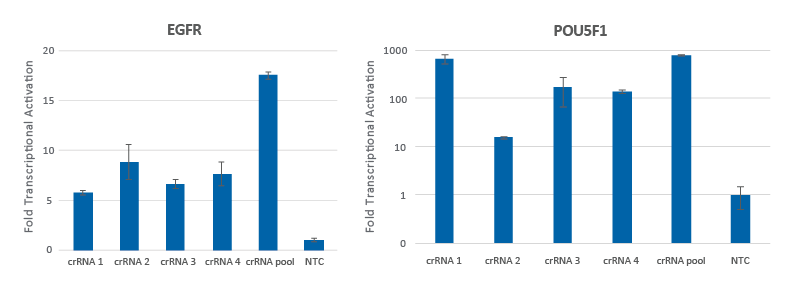
Individual CRISPRa crRNAs achieve robust target gene activation on their own, but when pooled together in a single reagent, enhanced activation levels can be achieved. U2OS cells stably expressing integrated CRISPRa dCas9-VPR were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA targeting EGFR or POU5F1. Four CRISPRa crRNAs were used either individually or pooled (to a total concentration of 25 nM). Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative fold transcriptional activation for each gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).
CRISPRa gene activation fold depends on the endogenous gene expression level
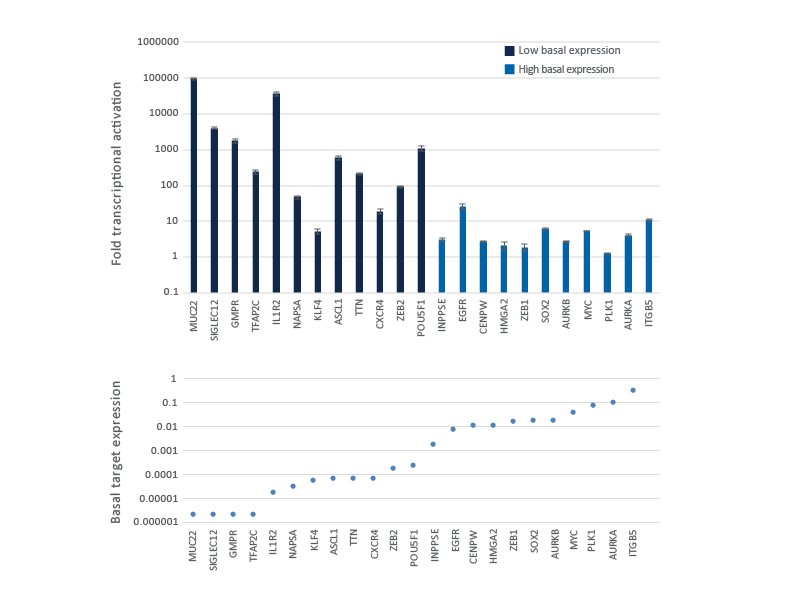
Genes with low or no basal expression are easier to activate to a robust level, while genes already expressed at high level are more difficult to further over-express. U2OS-dCas9-VPR stable cells were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA pools (25 nM) targeting 23 genes with different basal level of expression. Cells were harvested 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The CRISPRa-mediated fold transcriptional activation is shown in the upper graph where the genes are ordered from low to high level of basal transcript expression in samples treated with NTC control and is shown in the lower graph as basal target gene expression (compared to GAPDH control).
Immunofluorescence analysis demonstrates effective CRISPRa gene activation
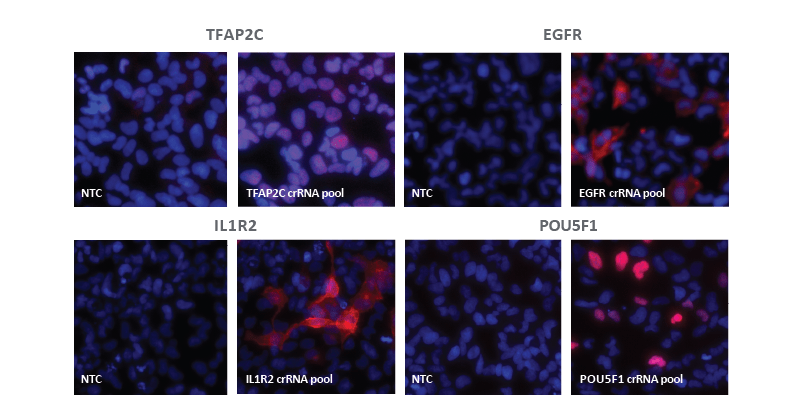
U2OS-dCas9-VPR stable cells were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent (0.2 µL/well) with CRISPRa synthetic crRNA:tracrRNA pool targeting TFAP2C, EGFR, IL1R2 or POU5F1 (25 nM total concentration) or NTC control. 72 hours post-transfection, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 followed by incubation with target specific primary antibodies and Dylight 550 conjugated secondary antibodies. Nuclei were stained with Hoescht 33342 Merged images are shown for cells treated with the target specific CRISPRa crRNA pools or NTC controls.
CRISPRa gene activation is observed at 24 hours and peaks at 48-72 hours
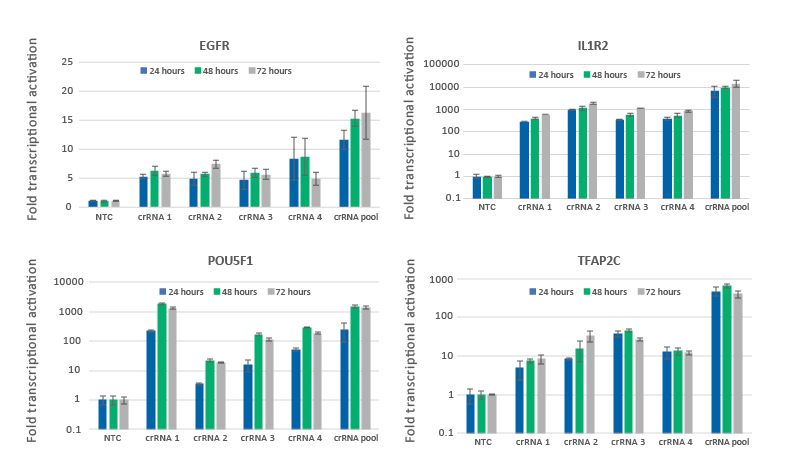
U2OS-dCas9-VPR stable cells were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA targeting EGFR, IL1R2, POU5F1 or TFAP2C. Four CRISPRa crRNAs were used either individually or pooled (to a total concentration of 25 nM). Cells were harvested at 24, 48, and 72 hours post-transfection and the relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
CRISPRa dose curve demonstrates highly effective gene activation at 25 nM working concentration
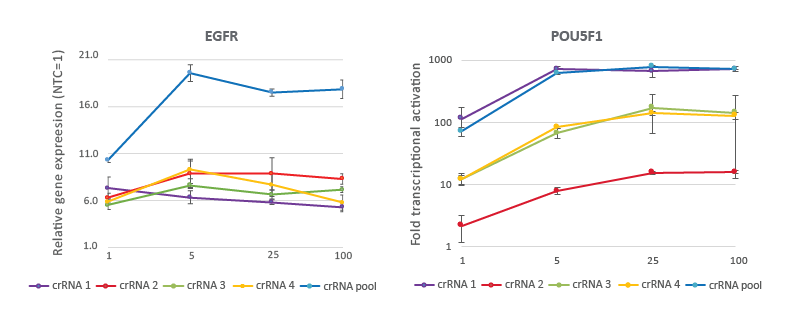
U2OS-dCas9-VPR stable cells were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with CRISPRa synthetic crRNA:tracrRNA targeting EGFR or POU5F1. The pre-designed crRNAs were used either individually or pooled at four concentrations (1, 5, 25, 100 nM). Cells were harvested 72 hours post-transfection and the relative gene expression was calculated using RT-qPCR. The relative expression of each gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control.
- Strezoska, Z., Dickerson, SM, et. al., CRISPR-mediated transcriptional activation with synthetic guide RNA. J Biotechnol. 2020 Aug 10;319:25-35. doi: 10.1016/j.jbiotec.2020.05.005. PubMed 32470463
-
Horlbeck MA, Gilbert LA, et. al., Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. 2016 Sep 23;5. pii: e19760. doi: 10.7554/eLife.19760. PubMed 27661255
-
Chavez A, Scheiman J et. al., Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015 Mar 2. doi: 10.1038/nmeth.3312. 10.1038/nmeth.3312 PubMed 25730490
-
Tanenbaum ME, Gilbert LA, et. al., A protein tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635-646. doi:10.1016/j.cell.2014.09.039. PubMed 25307933
Application notes
Posters
Protocols
Quick protocols
Safety data sheets
Related Products
Chemically synthesized trans-activating CRISPR RNA required for use with synthetic crRNA for fast and easy gene editing
Validated synthetic crRNA pool or individual for experimental optimization of transcription activation experiments.
Non-targeting controls to evaluate baseline cellular responses to CRISPRa components in the absence of gene target-specific crRNA.
Purified dCas9-VPR mRNA for co-transfection or electroporation with synthetic CRISPRa crRNA for gene activation
