- CRISPR activation reagents
- Strict-R inducible lentiviral dCas9-VPR CRISPRa
Strict-R Inducible CRISPRa Lentiviral System
How it works
System OFF: In the absence of doxycycline and Shield1, the system remains inactive. Any residual transcription from the TRE3G promoter produces degron-tagged dCas9-VPR, which undergoes rapid proteasomal degradation, minimizing unintended background activation.
System ON: Doxycycline administration activates the Tet-On 3G transactivator, inducing robust dCas9-VPR expression. Shield1 addition stabilizes the expressed dCas9-VPR protein, allowing accumulation and precise gene activation.
Schematic map of the Strict-R Inducible Lentiviral CRISPRa vector
Transcriptional and post-translational control with the Strict-R Inducible Lentiviral CRISPRa System

Diagram of the Strict-R Inducible Lentiviral CRISPRa System. In the absence of Doxycycline and Shield1, the system is “OFF”. Leaky bursts of transcription from the TRE3G promoter result in the translation of dCas9-VPR fused to a FKBP12-derived destabilizing domain (degron) that tags the protein for rapid proteasomal degradation . The addition of doxycycline induces potent transcription from the TRE3G promoter and the addition of Shield1 stabilizes dCas9-VPR thereby enabling robust target gene activation in the presence of a gene-specific sgRNA. Diagram created with BioRender.com.
Highlights
- Achieve reversible, small molecule-induced activation of target genes, providing tight regulation of gene expression.
- The dCas9-VPR fusion protein maximizes transcriptional activation with minimal off-target effects.
- The integration of the Tet degron system allows for rapid induction of dCas9-VPR expression upon the addition of doxycycline and Shield1, facilitating dynamic, time-controlled experiments.
- This system is expressed from a single lentiviral vector facilitating integration into your experimental workflow.
- Enables wide range of research applications including gene expression studies, functional genomics, and potential therapeutic applications.
- High quality, purified lentiviral particles for direct transduction with minimal cytotoxicity; delivered at titers of ≥ 1 x 107 TU/mL
- Options for either Blastacidin selection or EGFP fluorescent reporter.
Gene activation workflow using the Strict-R Inducible Lentiviral CRISPRa System

Figure 1: Gene overexpression workflow using the Strict-R Inducible Lentiviral CRISPRa System.
Tighter control of CRISPR activation with the Strict-R Inducible Lentiviral CRISPRa System
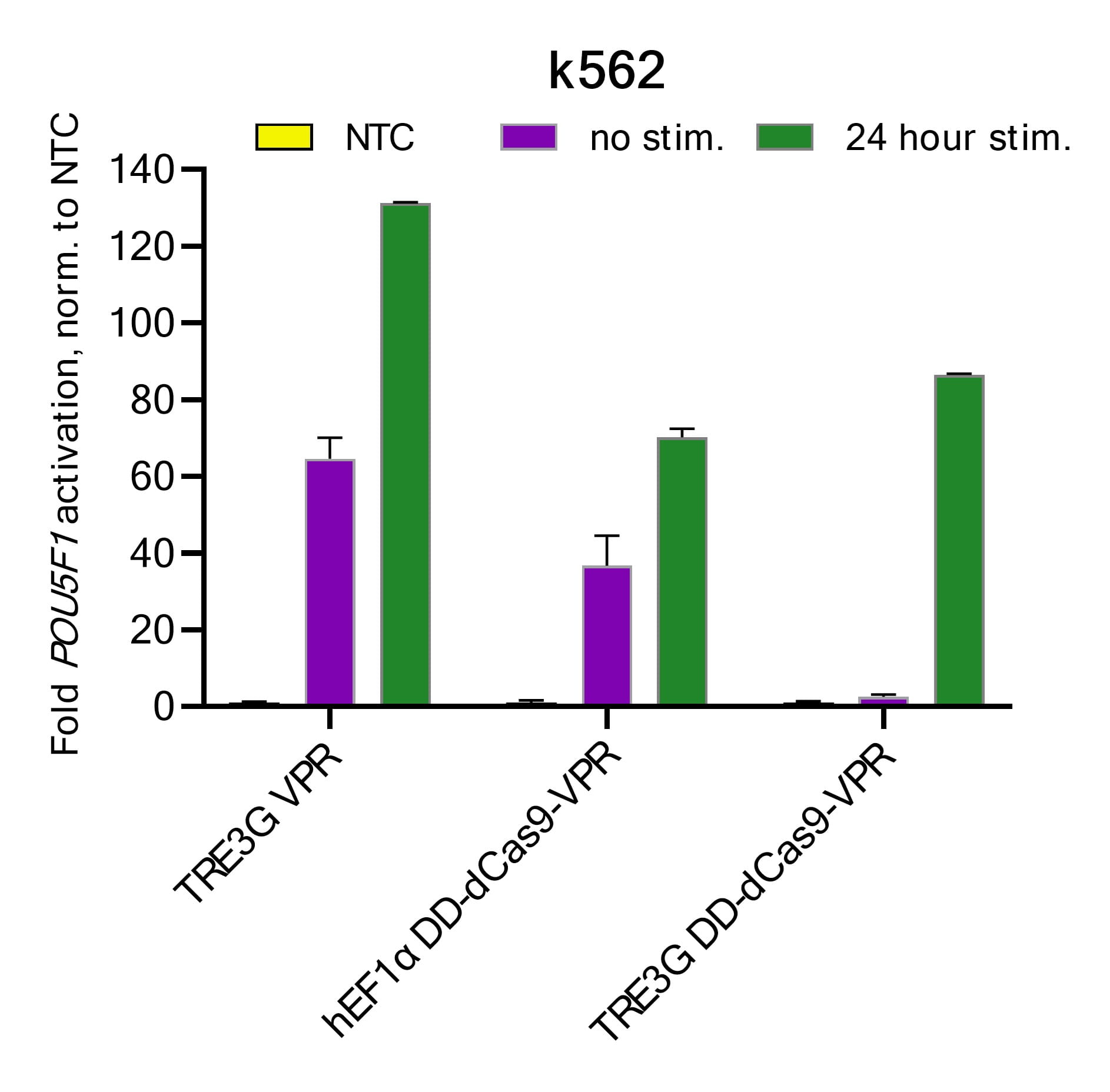
Figure 1: K562 cells were transduced with lentiviral particles containing the respective dCas9-VPR constructs at MOIs <0.3. Cells were selected with 10 µg/ mL blasticidin for two weeks with passaging every 3 to 4 days. After blasticidin selection, stable cell lines were transduced with either a CRISPRmod CRISPRa lentiviral sgRNA targeting POU5F1 or a CRISPRmod CRISPRa lentiviral non-targeting control (NTC) at MOIs of 0.3. After 48 hours, cells were selected with 2.5 µg/µL puromycin for 5 days. Cells were plated in 96-well plates at 20,000 cells/well and stimulated with 0.5 µg/ mL doxycycline and 0.25 nM Shield1 for 24 hours prior to harvest. Total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of POU5F1 was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. The combination of the TREG3G system and the FKBP12-derived destabilizing domain (DD) used in the Strict-R Inducible Lentiviral CRISPRa System significantly reduces background target gene activation (leakiness, purple) in the unstimulated state while enabling potent target activation with the addition of two highly cell-permeable small molecules.
Robust activation and minimal leakiness with the Strict-R Inducible Lentiviral CRISPRa System across cell lines and gene targets

Figure 2: U2OS, Hela, and human induced pluripotent stem cells (hiPSCs) expressing either the Strict-R Inducible Lentiviral CRISPRa System or dCas9-VPR under control of the constitutive hEF1α promoter (const. VPR, in black) were transduced with CRISPRmod™ CRISPRa lentiviral sgRNA targeting EGFR or ASCL1 or a CRISPRmod CRISPRa lentiviral non-targeting control (NTC) at MOIs of 0.3. After 48 hours, cells were selected with 2.5 µg/µL puromycin for 5 to 7 days. Cells were plated in 96-well plates at 20,000 cells/well and stimulated with 0.5 µg/ mL doxycycline and 0.25 nM Shield1 for 48 hours prior to harvest. Total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene upregulation was observed with the Strict-R Inducible Lentiviral CRISPRa System after 48 hours of stimulation, comparable to levels achieved with the constitutive dCas9-VPR system, with minimal background target gene activation (leakiness) observed in the unstimulated cell populations.
FACS can be used to select for Strict-R Inducible Lentiviral CRISPRa positive cells with high EGFP co-expression to enrich for CRISPRa activity in Jurkat cells

Figure 3: Jurkat cells were transduced with Strict-R Inducible Lentiviral CRISPRa EGFP particles at an MOI <0.3. Cells were expanded and then FACS was performed to sort the cells into two categories, GFP low and GFP hi (sic), based on fluorescence intensity. Following sorting, cells were replated in 6-well dishes and allowed to recover and expand. Cells were then transduced with CRISPRmod CRISPRa lentiviral sgRNA targeting EGFR or POU5F1 or a CRISPRmod CRISPRa lentiviral non-targeting control (NTC) at MOIs of 0.3. After 48 hours, cells were selected with 2.5 µg/µL puromycin for 5 days. Cells were plated in 96-well plates at 20,000 cells/well and stimulated with 0.5 µg/ mL doxycycline and 0.25 nM Shield1 for 48 hours prior to harvest. Total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Robust target gene upregulation was observed after 48 hours of stimulation with substantially more target activation in the GFP hi populations. Importantly, minimal background target gene activation (leakiness) was detected in both GFP low and GFP hi populations of unstimulated cells indicating enrichment for high effector expression does not significantly increase background activation.
Drug-induced activation of ASCL1 leads to upregulation of downstream targets DLL1 and DLL3 in hiPSCs

Figure 4: Human induced pluripotent stem cells (hiPSCs) expressing the Strict-R Inducible Lentiviral CRISPRa System were transduced with CRISPRmod CRISPRa lentiviral sgRNA targeting ASCL1 or a CRISPRmod CRISPRa lentiviral non-targeting control (NTC) at MOIs of 0.3. After 48 hours, cells were selected with 2.5 µg/µL puromycin for 7 days. Cells were plated in 96-well plates at 20,000 cells/well and stimulated with 0.5 µg/ mL doxycycline and 0.25 nM Shield1 for 48 hours prior to harvest. Total RNA was isolated and the relative gene expression was measured using RT-qPCR for ASCL1 and its downstream Delta gene targets DLL1 and DLL3. The relative expression of each gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to an NTC. Activation of Delta genes by proneural transcription factors such as ASCL1 is an evolutionarily conserved step in neurogenesis that results in activation of Notch signaling and maintenance of an undifferentiated state in a subset of neural progenitors. Stimulation with doxycycline and Shield1 induced potent activation of ASCL1 which resulted in marked upregulation of both DLL3 and DLL1. Crucially, low level background activation of ASCL1 did not significantly impact DLL1 or DLL3 expression.
- Bhaya, et al., CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273-297 (2011).
- M. Jinek, et al., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816-821 (2012).
- L.S. Qi, et al., Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152, 1173-83 (2013).
- L.A.Gilbert, et al., CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 154, 442-51 (2013).
- A.W. Cheng, et al., Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res., 23, 1163-71 (2013).
- L. A. Gilbert, et al., Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159, 647–661 (2014).
- M. E. Tanenbaum, et al., A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 159, 635–646 (2014).
- S. Konermann, et al., Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517, 583–588 (2015).
- R. Loew, et al., Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 10, 81 (2010).
- Banaszynski, Laura A et al., A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 126, 995-1004 (2006).
- Egeler, Emily L et al., Ligand-switchable substrates for a ubiquitin-proteasome system. The Journal of Biological Chemistry. 286, 31328-36 (2011).
- M. A. Horlbeck et al., Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760 (2016).
- Maynard-Smith, Lystranne A et al., A directed approach for engineering conditional protein stability using biologically silent small molecules. The Journal of Biological Chemistry. 282, 24866-72 (2007).
LentiBOOST Lentivirus Transduction Enhancer is a uniquely formulated transduction reagent that can be used with or without lentivirus spinfection in order to increase successful viral transduction events while preserving cell viability. Especially critical for preserving precious primary cells from patient cohorts, or, for engineering complex animal models; improving transduction efficiency can save time and costs by increasing the success of each editing/transduction step, or, even avoid the loss of irreplaceable samples. Additionally, LentiBOOST technology is already used in the manufacturing of a number of clinical stage therapies providing the opportunity to demonstrate improved workflow applicability to the clinic.
LentiBOOST can be purchased through the Dharmacon Reagents catalog.
To learn more about LentiBOOST technology visit the Revvity LentiBOOST webpage.
Supporting Data
Improved CD8+ T-cell SMARTvector™ shRNA lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
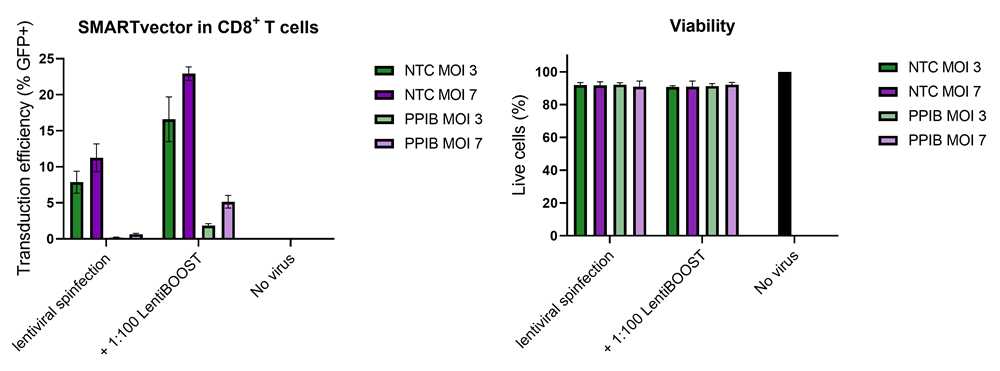
100,000 primary human CD8+ T cells were transduced with either 30,000 (MOI 3, green) or 70,000 (MOI 7, purple) TUs of SMARTVector™ mCMV tGFP Lentiviral Control Particles targeting either NTC or PPIB along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by a four hour incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency (%GFP+ out of live cells) and viability were determined 5 days post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved CD4+ and CD8+ T-cell Edit-R™ All-in-one sgRNA/Cas9 lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer
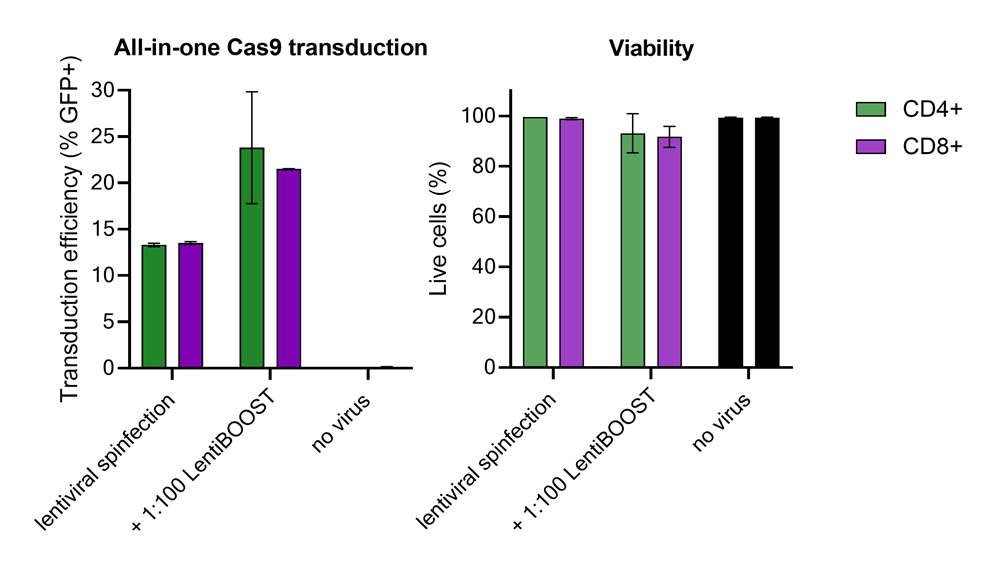
100,000 primary human CD4+ and CD8+ T cells from two donors were transduced with 250,000 TUs of Edit-R GFP Delivery controls mCMV along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST technology markedly improved transduction efficiencies without significantly impacting cell viability.
Improved transduction of human induced pluripotent stem cells (hiPSCs) with the Strict-R™ Inducible CRISPRa lentiviral system transduction using LentiBOOST™ Lentivirus Transduction Enhancer

10,000 WTC hiPS cells were transduced with either 20,000 (MOI 2, green) or 40,000 (MOI 4, purple) TUs of Strict-R™ Inducible EGFP dCas9-VPR Lentiviral Particles along with 1:100 LentiBOOST transduction enhancer. Cells were centrifuged at 800 x g for one hour at 32 °C followed by an overnight incubation prior to removal of lentiviral particles and transduction enhancer. Transduction efficiency and viability were determined 72 hours post-transduction by flow cytometry. The addition of LentiBOOST markedly improved transduction efficiencies without significantly impacting cell viability.
Safety data sheets
Related Products
Predesigned CRISPRa lentiviral sgRNAs for highly efficient gene activation in human and mouse models; available as glycerol stocks and high-titer purified particles.
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.
Lentiviral sgRNA constructs bioinformatically designed and validated to not target any gene in human or mouse genomes.
Validated CRISPRa lentiviral sgRNA positive controls for optimization of experimental conditions and quantification of gene activation efficiency.

