- CRISPR interference reagents
- dCas9-SALL1-SDS3 mRNA
CRISPRmod CRISPRi dCas9-SALL1-SDS3 mRNA
A lentiviral-free option for dCas9-SALL1-SDS3 expression
Purified dCas9-SALL1-SDS3 mRNA for co-transfection or electroporation with synthetic CRISPRi sgRNA for repression of targeted gene transcription

CRISPRi dCas9-SALL1-SDS3 mRNA expresses a human codon-optimized version of the nuclease-deactivated S. pyogenes Cas9 gene, fused to our proprietary transcriptional repressors (SALL1 and SDS3). When paired with a well-designed guide RNA that targets a gene near the native transcription start site, expression is repressed.

Review our CRISPRi applications page to get an overview of the technology!
Highlights of using CRISPRi dCas9-SALL1-SDS3 mRNA
- No need to generate stable dCas9-SALL1-SDS3-expressing cell lines
- Simply co-transfect CRISPRi synthetic sgRNA and dCas9-SALL1-SDS3 mRNA using DharmaFECT Duo transfection reagent or co-electroporate into cells.
- Co-expression of either EGFP or puromycin resistance allows for easy transfection optimization or enrichment using FACS or antibiotic selection.
- No need for optimal promotor selection, as mRNA is ready for translation
Requirements for a CRISPRi experiment using dCas9-SALL1-SDS3 mRNA
- CRISPRi dCas9-SALL1-SDS3 mRNA
- CRISPRi synthetic sgRNA for your target gene (see Workflow tab)
CRISPRi workflow using synthetic sgRNA and dCas9-SALL1-SDS3 mRNA
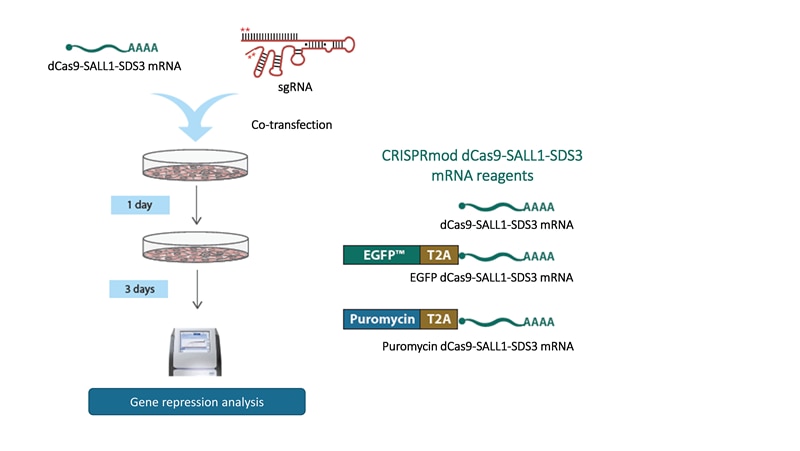
Co-transfect or electroporate cells with CRISPRi synthetic sgRNA and CRISPRi dCas9-SALL1-SDS3 mRNA. Then enrich cell populations using fluorescence or puromycin resistance options. This system is best suited for rapid, transient gene repression studies.
Robust, transient gene repression with CRISPRi dCas9-SALL1-SDS3 mRNA and CRISPRi synthetic sgRNA

K562 and Jurkat cells were nucleofected with dCas9-KRAB or dCas9-SALL1-SDS3 mRNA (2 µg), and pooled CRISPRi synthetic sgRNAs (5 µM) targeting PPIB and SEL1L via a Lonza 96-well Shuttle system. WTC-11 hiPS cells were nucleofected with dCas9-KRAB or dCas9-SALL1-SDS3 mRNA (1 µg) and pooled CRISPRi synthetic sgRNA (3 µM) targeting PPPIB or SEL1L via a Lonza 96-well Shuttle system. Cells were harvested 72 hours post-nucleofection. Total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative gene expression for each target gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).
Co-transfection of dCas9-SALL1-SDS3 mRNA and CRISPRi synthetic sgRNA provides greater gene repression than transfection into a stable dCas9-SALL1-SDS3 expressing cell line
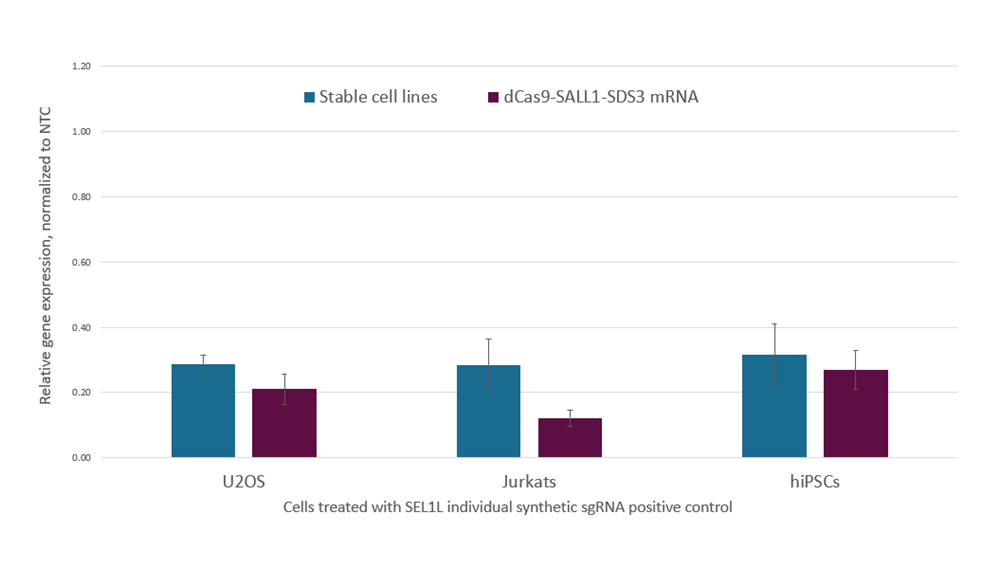
Data was compiled from multiple experiments and matched for the time point of analysis and delivery method. U2OS cells stably expressing integrated dCas9-SALL1-SDS3 were plated at 10,000 cells/well and transfected using DharmaFECT 4 Transfection Reagent with synthetic sgRNA (25 nM) targeting SEL1L, parental U2OS cells were plated at 10,000 cells/well and co-transfected using DharmaFECT Duo Transfection Reagent with dCas9-SALL1-SDS3 mRNA (200 ng) and CRISPRi synthetic sgRNA (25 nM) targeting SEL1L. Jurkat cells stably expressing integrated dCas9-SALL1-SDS3 were nucleofected with CRISPRi synthetic sgRNA (5 µM) targeting SEL1L, parental Jurkats cells were nucleofected with dCas9-SALL1-SDS3 mRNA (2 µg), and CRISPRi synthetic sgRNA (5 µM) targeting SEL1L via a Lonza 96-well Shuttle system. WTC-11 human iPS cells stably expressing integrated dCas9-SALL1-SDS3 were nucleofected with CRISPRi synthetic sgRNA (3 µM) targeting SEL1L, parental WTC-11 hiPS cells were nucleofected with dCas9-SALL1-SDS3 mRNA (1 µg) and CRISPRi synthetic sgRNA (3 µM) targeting SEL1L via a Lonza 96-well Shuttle system. U2OS cells were harvested 48 hours post-transfection, Jurkat and hiPS cells were harvested 72 hours post-nucleofection. Total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative gene expression for SEL1L was calculated with the Cq method using GAPDH or ACTB as the housekeeping gene and normalized to a non-targeting control (NTC).
CRISPRi dCas9-SALL1-SDS3 mRNA co-expressing PuroR allows for enrichment using puromycin selection
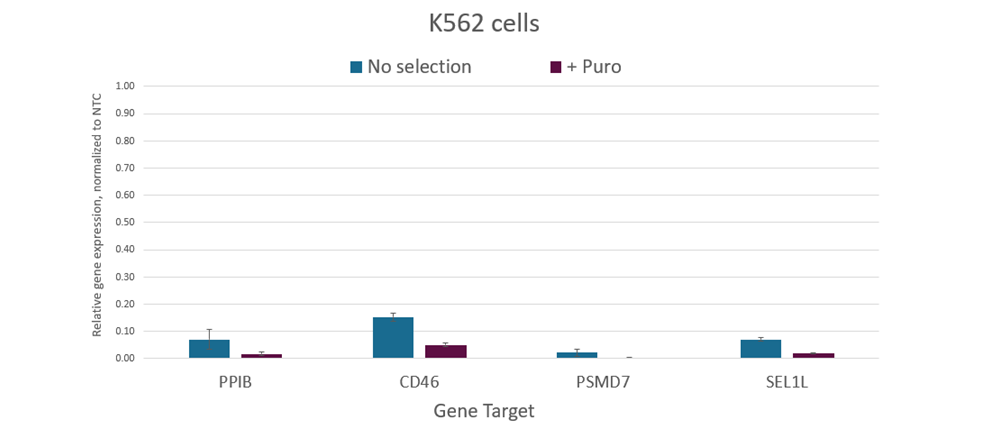
K562 cells were nucleofected with Puro dCas9-SALL1-SDS3 mRNA (2 µg), and pooled CRISPRi sgRNA targeting BRCA1, PPIB, CD46, PSMD7, SEL1L, or ST3GAL4 via a Lonza 96-well Shuttle system. At 24 hours post-nucleofection 2 µg/ml of puromycin growth media was added to the cells and a duplicate plate received normal growth media. At 48 hours post-nucleofection total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each target gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).WTC-11 hiPS cells were plated at 5,000 cells per well in clear 96-well plates. After 24 hours, cells were co-transfected with CRISPRi PuroR dCas9-SALL1-SDS3 mRNA (200 ng) and pooled CRISPRi synthetic sgRNA (25nM) targeting either PPIB or ST3GAL4 using DharmaFECT Duo Transfection Reagent. At 16 hours post-transfection, 2 µg/ml of puromycin growth media was added to the cells and a duplicate plate received normal growth media. At 24 hours post-transfection the puromycin containing growth media was removed from the cells and replaced with antibiotic-free growth media. At 72 hours post-transfection total RNA was isolated and relative gene expression was measured using RT-qPCR. The relative expression of each target gene was calculated with the Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control (NTC).
CRISPRi dCas9-SALL1-SDS3 mRNA co-expressing EGFP allows for FACS enrichment
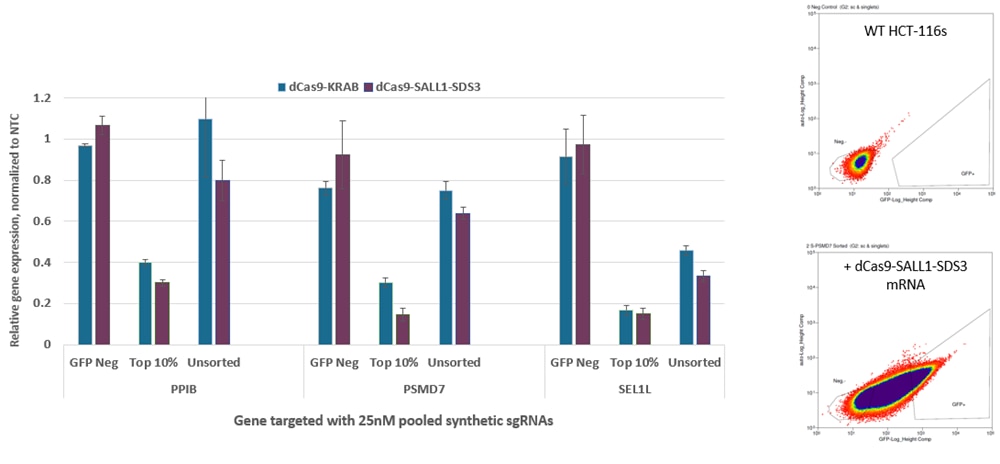
HCT-116 cells were plated at 400,000 cells per well in clear 6-well plates. After 24 hours, cells were co-transfected with EGFP dCas9-KRAB or EGFP dCas9-SALL1-SDS3 mRNA (2.5 µg), and pooled CRISPRi synthetic sgRNA targeting PPIB, PSMD7, or SEL1L (25 nM) using DharmaFECT Duo Transfection Reagent. At 24 hours post-transfection cells were trypsinized and FACS was performed to sort the cells into two categories: GFP negative (GFP Neg), and top 10% GFP expressing (Top 10%). Following sorting, cells were replated in 6-well dishes and allowed to recover. Cells were harvested after 24 hours of recovery, total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative expression of each target gene was calculated with the Cq method using GAPDH as the housekeeping gene and normalized to a non-targeting control (NTC).
Protocols
-
CRISPRmod dCas9-VPR or dCas9- SALL1-SDS3 mRNA and synthetic guide RNA transfection protocol
-
Dose response curve for antibiotic selection of mammalian cells (kill curve) - Protocol
-
Electroporation of CRISPRmod dCas9-VPR or dCas9-SALL1-SDS3 mRNA and synthetic guide RNA for gene modulation
-
Enrichment of transfected cells with CRISPRmod EGFP dCas9-VPR or dCas9-SALL1-SDS3 mRNA
Safety data sheets
Technical manuals
Related Products
Validated CRISPRi synthetic pools or individual sgRNA for optimization of transcriptional repression experiments.
Validated CRISPRi synthetic pools or individual sgRNA for evaluation of transcriptional repression experiments.
LentiBOOST transduction enhancer can increase successful viral transduction in challenging to transduce cells, or, complex cellular engineering work; while preserving cell viability and minimizing the amount of viral particles required for your experiment. LentiBOOST technology is actively used in the production of clinical stage lentivirally delivered therapies, including some approved therapies, providing a direct path to therapeutic applicability for your research studies. Tested with Dharmacon Lentiviral reagents.
Perform repression of thousands of genes at the transcriptional level with a single transduction of CRISPRi all-in-one pooled lentiviral library eliminating the need to perform sequential transductions.
