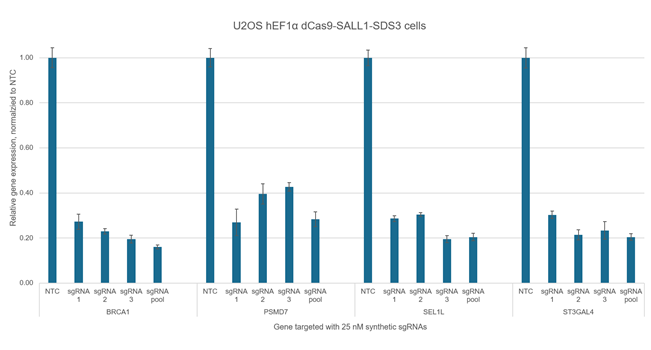
CRISPRmod CRISPRi synthetic sgRNA non-targeting controls
Validated CRISPRi synthetic pools or individual sgRNA for evaluation of transcriptional repression experiments
Used to evaluate baseline cellular responses to CRISPRi components in the absence of gene target-specific sgRNA