- Gene editing
- Gene editing reagents
- Edit-R HDR Plasmid Donor Kits
Edit-R HDR plasmid donor kits
Kit for assembly of a plasmid DNA EGFP, mKate2, or custom sequence HDR knock-in donor

Gene engineering using CRISPR-Cas9 relies on two pathways for repair of the cut target DNA, non-homologous end joining (NHEJ) and homology-directed repair (HDR). Precise repair of the DNA to generate a specific mutation or insert or remove a desired sequence requires the HDR pathway. This process is dependent on the availability of a DNA donor template that can be used to repair the double-strand break in the DNA. One of the most common applications is insertion of a fluorescent reporter sequence on one end of a gene. The Edit-R HDR Plasmid Donor Kit enables rapid and easy creation of a plasmid donor for insertion of the EGPF or mKate2 fluorescence reporter or a user-provided custom sequence into almost any genomic location.
Highlights
- Fast and efficient assembly of a DNA donor plasmid
- Validated protocols for design, assembly, and confirmation of the donor plasmid
- Utilize the CRISPR Design Tool for designing the crRNA to the cut site
- Seamless integration with the HDR Donor Designer for designing the primers to create the required 5′ and 3′ homology arms flanking the insertion site
The HDR plasmid donor kit contains all the DNA components to build and quality check a custom DNA donor template plasmid to introduce a fluorescent reporter to almost any genomic location when coupled with Edit-R HDR DNA donor plasmid homology arm custom primer pairs.
The HDR Donor Designer is used to design and order the PCR primers for the amplification of the 5′ and 3′ homology arms, required for assembly of the plasmid.
Kit Components & Amounts
| Cat # | Description | Item# | Amount |
|---|---|---|---|
| UK-008100-01-10 | Edit-R HDR Plasmid Donor Kit - EGFP | ||
| Edit-R HDR Plasmid Donor Backbone | U-008300-01-1200 | 1200 ng | |
| Edit-R HDR insert - EGFP | U-008100-01-250 | 250 ng | |
| Edit-R Colony PCR Primer EGFP Forward | U-008100-FB-05 | 5 nmol | |
| Edit-R Colony PCR Primer EGFP Reverse | U-008100-RA-05 | 5 nmol | |
| Edit-R Colony PCR Primer Backbone Forward | U-008300-FW-05 | 5 nmol | |
| Edit-R Colony PCR Primer Backbone Reverse | U-008300-RV-05 | 5 nmol | |
| UK-008200-03-10 | Edit-R HDR Plasmid Donor Kit -mKate2 | ||
| Edit-R HDR Plasmid Donor Backbone | U-008300-01-1200 | 1200 ng | |
| Edit-R HDR insert - mKate2 | U-008200-01-250 | 250 ng | |
| Edit-R Colony PCR Primer mKate2 Forward B | U-008200-FB-05 | 5 nmol | |
| Edit-R Colony PCR Primer mKate2 Reverse A | U-008200-RA-05 | 5 nmol | |
| Edit-R Colony PCR Primer Backbone Forward | U-008300-FW-05 | 5 nmol | |
| Edit-R Colony PCR Primer Backbone Reverse | U-008300-RV-05 | 5 nmol | |
| UK-008300-01-10 | Edit-R HDR Plasmid Donor Kit - Universal | ||
| Edit-R HDR Plasmid Donor Backbone | U-008300-01-1200 | 1200 ng | |
| Edit-R Colony PCR Primer Backbone Forward | U-008300-FW-05 | 5 nmol | |
| Edit-R Colony PCR Primer Backbone Reverse | U-008300-RV-05 | 5 nmol |
Diagram of the plasmid donor cloning workflow
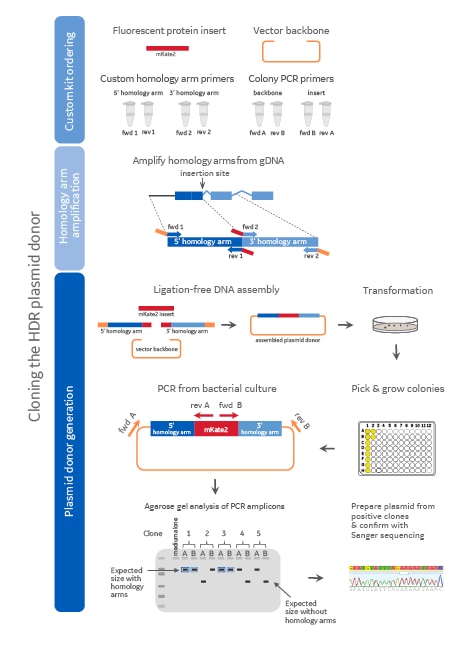
Diagram of the plasmid donor cloning workflow including component ordering, homology arm amplification, and plasmid donor generation. Colors on the diagram indicate the origin of the DNA (dark blue = 5' homology arm, light blue = 3' homology arm, red = mKate2 fluorescent reporter sequence, orange = plasmid backbone). medium alone = media-only negative PCR control.
LMNA tagged at the N-terminus with EGFP in U2OS cells
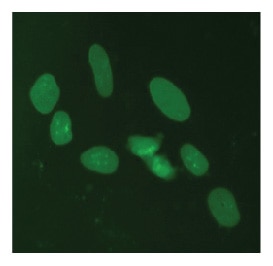
U2OS cells were transfected with DharmaFECT Duo, Cas9 mRNA, synthetic crRNA-tracrRNA targeting LMNA, and an EGFP donor plasmid specific to the N-terminus of LMNA containing ~1000 bps of homology per homology arm. Cells were passaged as needed, and imaged 7-days post transfection on an InCell 2200 high content fluorescent microscope.
mKate2-SEC61B fusion in U2OS cells
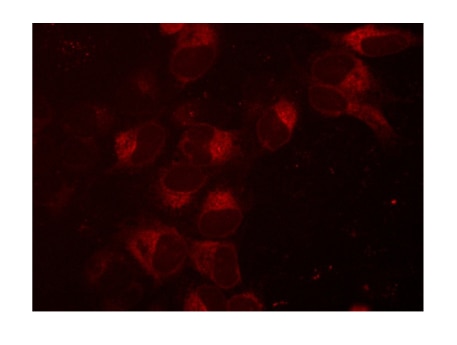
Creation of an mKate2 N-terminal fusion to the SEC61B gene, a component of protein transport in the endoplasmic reticulum. U2OS cells were transfected with 50 nM SEC61B-crRNA:tracrRNA, 25 nM Cas9 protein, 2 ng/µL SEC61B mKate2 HDR donor plasmid, and 0.3 µL/well DharmaFECT Duo and visualized on a fluorescent microscope seven days post-transfection. Note: Ribonucleoprotein(RNP) complex ratio is 2:1 for Cas9 protein transfection.
U2OS cell line with EGFP-LMNA and mKate2-SEC61B fluorescent reporters generated through multiplexed CRISPR-Cas9-mediated knock-in
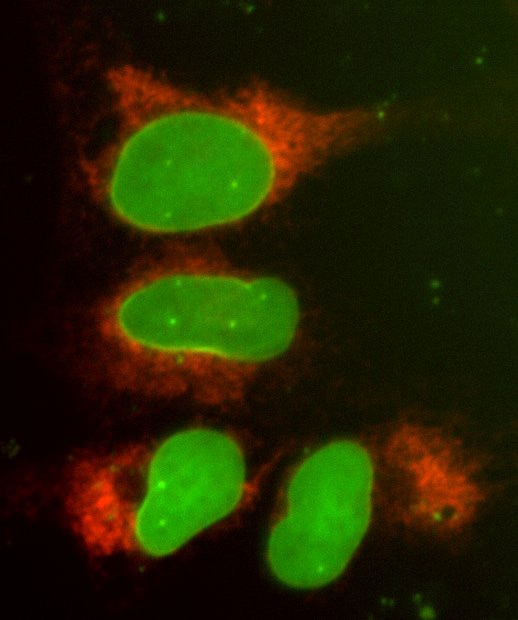
Briefly, synthetic crRNAs targeting LMNA and SEC61B were designed to generate double-strand breaks near the N-termini of each of the proteins using the CRISPR RNA Configurator. The Edit-R HDR Donor Designer was used to design reagents to generate EGFP and mKate2 donor plasmids specific to the N-termini of LMNA and mKate2, respectively. Both donor plasmids contained ~1000 base pairs of homology per homology arm. U2OS cells were plated at 10,000 cells per well in a 96-well plate the day before transfection. Cells were transfected using 0.3 µL/well DharmaFECT Duo transfection reagent according to optimized conditions determined in previous experiments. The following reagents were transfected into the cells simultaneously: 200 ng Edit-R™ Cas9 Nuclease mRNA, 25 nM synthetic crRNA:tracrRNA targeting LMNA, 25 nM synthetic crRNA:tracrRNA targeting SEC61B, 200ng EGFP-LMNA donor plasmid and 200 ng mKate2-SEC61B donor plasmid. Cells were passaged three times and imaged 7-days post transfection on a Nikon inverted epifluorescent microscope to identify single cells that contained both EGFP (green) and mKate2 (red) insertion.
LMNA tagged at the N-terminus with EGFP and SEC61B tagged at the N-terminus with mKate2 in U2OS cells
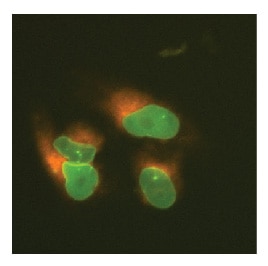
U2OS cells were transfected with DharmaFECT Duo, Cas9 mRNA, synthetic crRNA-tracrRNA targeting LMNA, synthetic crRNA-tracrRNA targeting SEC61B, an EGFP donor plasmid specific to the N-terminus of LMNA containing ~1000 bps of homology per homology arm, and a mKate2 donor plasmid specific to the N-terminus of SEC61B containing ~1000 bps of homology per homology arm. Cells were passaged as needed, and imaged 7-days post transfection on an InCell 2200 high content fluorescent microscope to identify single cells that contained both EGFP and mKate2 insertion.
Dharmacon™ Edit-R™ mKate2 knock-in donor
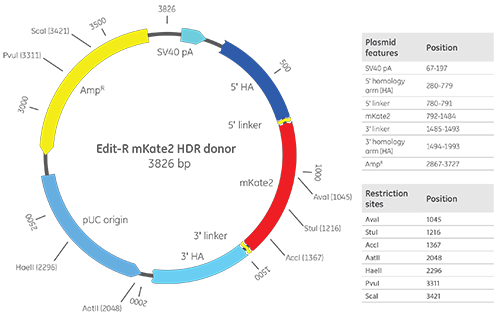
Edit-R mKate2 plasmid donor map with important vector features. The example includes 500 bp homology arms for reference.
Detection of successful cloning by colony PCR
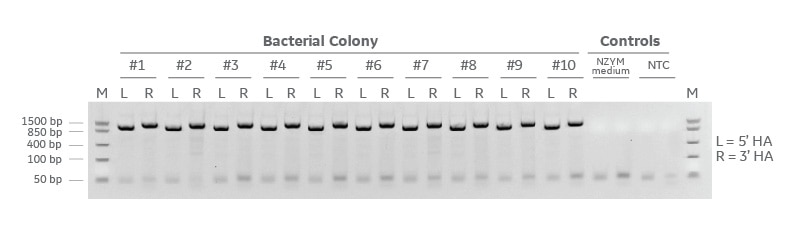
Gibson cloning results of the Edit-R donor plasmid kit with universal backbone, EGFP insert, and two PCR generated homology arms. The resulting cloning reaction was transformed in NEB 5-alpha chemically competent cells and grown at 37 degrees C overnight. 10 bacterial colonies were selected at random and colony PCR was performed to detect the presence of each homology arm (L=5’ homology arm and R=3’ homology arm). Colony PCR amplicons were run on a 2% agarose E-gel containing ethidium Bromide and a FastRuler low molecular weight molecular weight marker.
Workflow to acquire mKate2 knock-in clonal cell line
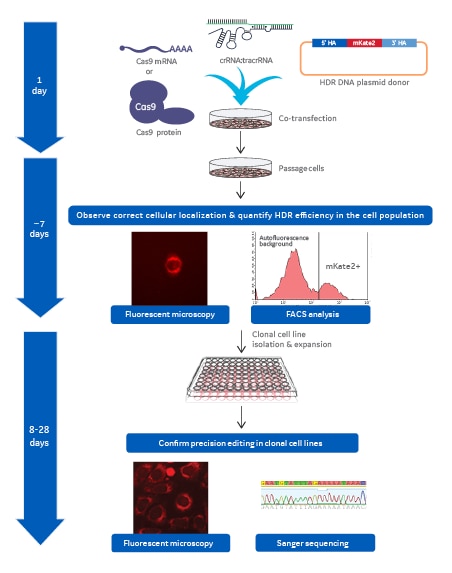
Workflow showing the major process steps and general timing necessary to generate a mKate2 knock-in clonal cell line.
