- Gene editing
- Pin-point™ Reagents
- Pin-point™ Base Editing mRNA
Pin-point™ Base Editing mRNA
|
Pin-point™ base editing mRNAs harness the DNA targeting ability of a nickase Cas9, with the DNA editing activity of a cytidine deaminase to introduce targeted point mutations when used in combination with Pin-point synthetic sgRNA. The platform consists of an mRNA for translation of a mammalian codon-optimized nCas9 and an mRNA for translation of a mammalian codon-optimized deaminase fused to an aptamer binding protein. Pin-point nCas9 mRNA The Pin-point nCas9 mRNA contains a human codon-optimized version of the S. pyogenes nickase Cas9 (D10A) with a 5’ and 3’ nuclear localization signal (NLS), fused to UGI. The nCas9 mRNA is in vitro transcribed, capped using CleanCap AG, and polyadenylated for translation and nuclear localization of the protein. Pin-point Rat APOBEC mRNA The Pin-point Rat APOBEC mRNA contains a human codon-optimized version of an aptamer binding protein fused to a cytidine deaminase base editor enzyme with a 5’ NLS. The Rat APOBEC mRNA is in vitro transcribed, capped using CleanCap AG, and polyadenylated for translation and nuclear localization of the protein. |
128 nt Pin-point sgRNA are available now
|
Modified or unmodified, which one should I use?
Both Pin-point nCas9 mRNA and Pin-point Rat APOBEC mRNA come in either unmodified or fully substituted with 5-methoxy-uridine (5moU modified) formats for optimized usage in different cell types. Additionally, each experiment with the Pin-point base editing system requires usage of nCas9 and Rat APOBEC mRNAs in combination with Pin-point synthetic sgRNA. View the "Ordering information" tab to find a table of the suggested use of the Pin-point mRNA formulations by cell type. Read our application note that discusses specific use cases for selecting unmodified versus 5moU modified mRNAs.
Base editing with the Pin-point platform induces specific nucleotide changes without the formation of DNA double-strand breaks or indels.
This system consists of three components:
- a nuclease-defective “nickase” nCas9 fused to a uracil glycosylase (UGI) inhibitor that only cuts or “nicks” a single strand of DNA (Komor, 2016).
- a cytidine deaminase base editor (Rat APOBEC) fused to an aptamer binding protein. The cytidine deaminase enzyme converts C-G base pairs to T-A base pairs if the base pairs are within the editing window, (positions 4 to 8 at the PAM-distal end of the protospacer) (Collantes, 2021).
- an aptameric single guide RNA (sgRNA) that recruits the nCas9 and the aptamer-deaminase fusion to a specific DNA target site.
Delivery of all three components into mammalian cells induces C-G to T-A base conversion at the targeted site. This system can be used to make gene knockouts (through the introduction of premature stop codons (Billon, 2017), the disruption of splice donor and splice acceptor sites (Webber, 2019)), or to introduce point mutations.
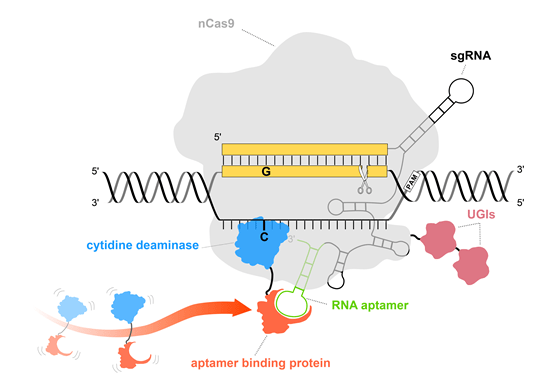
Illustration of Pin-point base editing system.
The utilization of nCas9 (light gray) ensures that no DNA double-strand breaks occur and DNA damage response pathway is not triggered. Pin-point sgRNA contains an aptamer (green) that is used to recruit deaminase (blue) via aptamer binding protein (orange) to perform base editing.
TriLink BioTechnologies LLC is the owner of the CleanCap® Technology. “CleanCap®”, “TriLink®” and “TriLink Biotechnologies®” are registered trademarks of TriLink BioTechnologies LLC.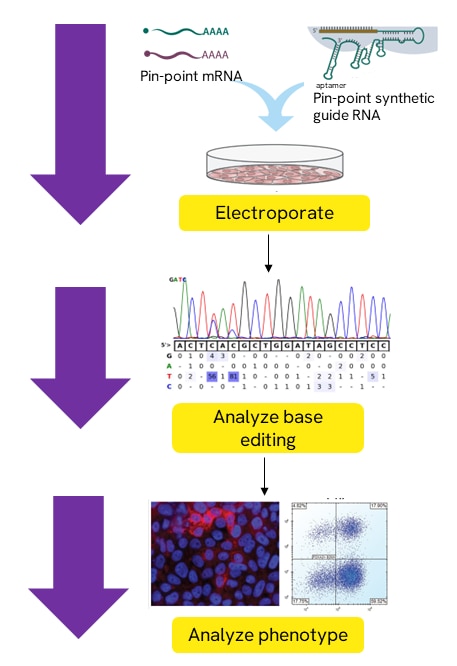
Base editing with Pin-point nCas9 mRNA, Pin-point Rat APOBEC mRNA, and Pin-point synthetic sgRNA.
All three components are electroporated into a specific cells type of interest. Base editing may then be observed using analysis primers to amplify and sequence the base editing region of interest. A phenotype may be analyzed to assess target protein knockout.
Table 1: Should I select Pin-point unmodified or Pin-point 5moU modified mRNAs?
|
Suggested use of Pin-point mRNAs by cell type |
|
|---|---|
| HEK293T cells | unmodified |
| Primary human T cells | unmodified |
| Induced pluripotent stem cells (iPSCs) | unmodified |
| Hemopoietic Stem and Progenitor Cells (HSPCs) | 5moU modified |
Table 2: How much Pin-point mRNA do I need?
|
Number of reactions by well/format size* |
|||||
|---|---|---|---|---|---|
| HEK 293T cells (50,000 cells per well in a 96 well plate) | T cells (250,000 cells per well in a 24 well plate) | ||||
| Pin-point nCas9 mRNA | Pin-point Rat APOBEC mRNA | Pin-point nCas9 mRNA | Pin-point Rat APOBEC mRNA | ||
| 1 µg/well | 0.1 µg/well | 1.56 µg/well | 0.22 µg/well | ||
| 20 µg (2 µg/µl) | 20 | 200 | 12 | 90 | |
| 100 µg (2 µg/µl) | 100 | 1000 | 64 | 450 | |
| 500 µg (2 µg/µl) | 500 | 5000 | 320 | 2252 | |
Table 3: How much Pin-point sgRNA do I need?
| Number of reactions by well/format size* | ||||
|---|---|---|---|---|
| HEK 293T cells (50,000 cells per well in a 96 well plate) | T cells (250,000 cells per well in a 24 well plate) | |||
| 60 pmol/well | Singleplex 20 pmol/well or Multiplex (3 guides) at 20 pmol/well each |
|||
| 2 nmol | 33 | 100 | ||
| 5 nmol | 83 | 250 | ||
| 10 nmol | 166 | 500 | ||
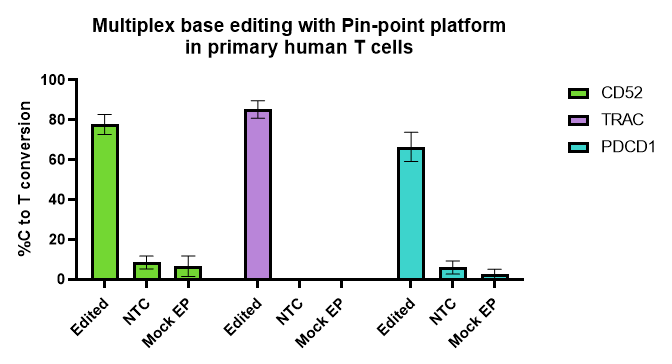
Figure 1
Pin-point base editing platform is efficient and reproducible across many experiments and T cells donors.
Activated T cells were electroporated with Pin-point mRNAs and Pin-point synthetic sgRNAs for the 3 targets (CD52, PDCD1 and TRAC), or non-targeting control (NTC #1) sgRNA. Mock-electroporated (EP) cells were used as negative controls. Cells were harvested 6-7 days post electroporation, and base editing levels were calculated from Sanger sequencing data of PCR products, using the Pin-point analysis primers. N = 8 T cell donors, 5 independent experiments with duplicate or triplicate technical replicates. Bars are averages ± SD.
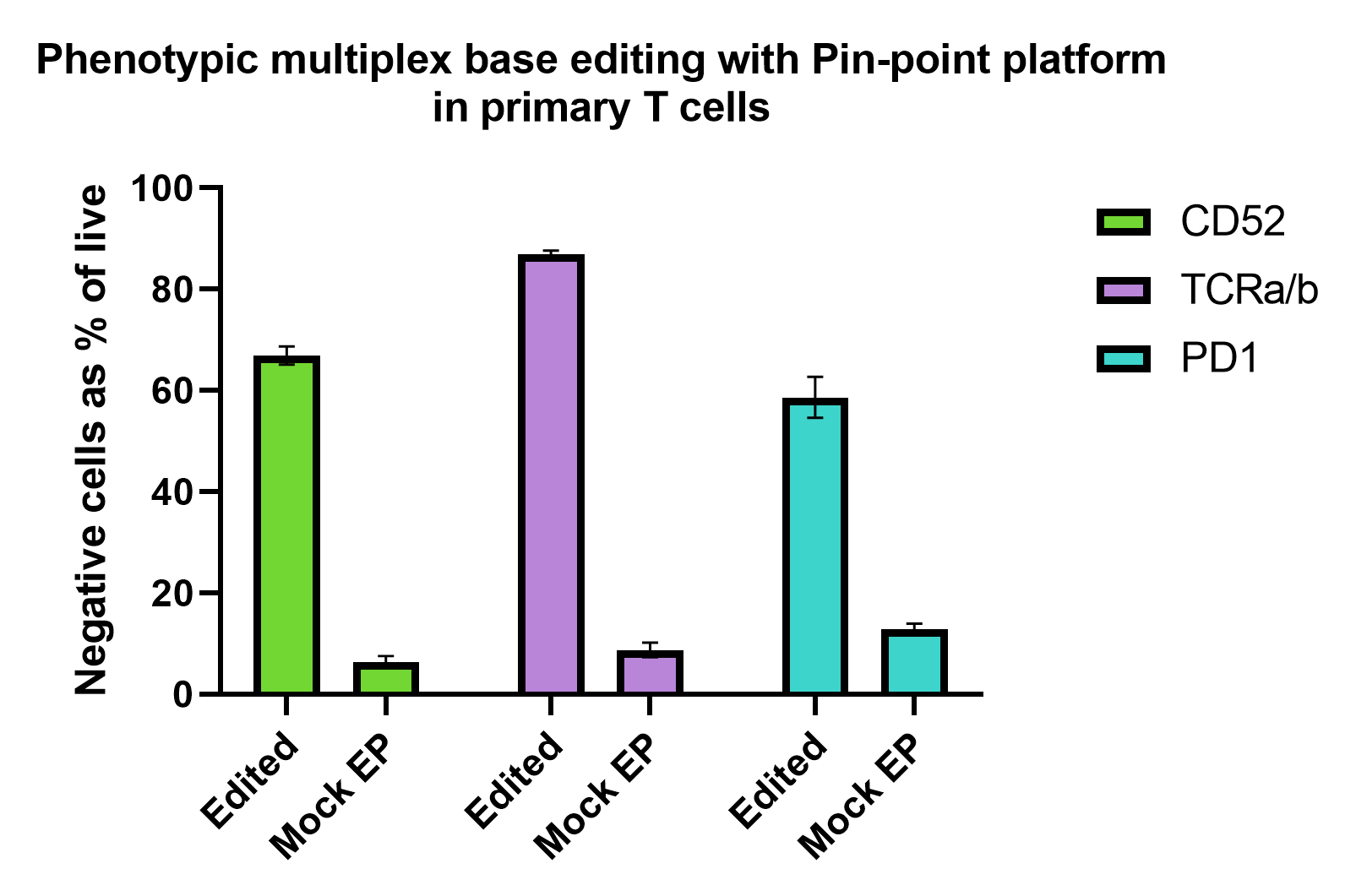
Figure 2
Pin-point base editing platform efficiently induces multiplex gene knockout in primary T cells. Activated T cells were electroporated with Pin-point mRNAs and Pin-point synthetic sgRNAs for the 3 targets (CD52, PDCD1 and TRAC) and analyzed by flow cytometry 7 days post electroporation. To induce the expression of PD1, T cells were cultured in the presence of phorbol 12- myristate 13-acetate and ionomycin for 48 h prior. Expression of CD52 and TCRa/b were analysed on non-stimulated cells. Percentage of negative cells for each marker in the edited and mock-electroporated (EP) reported as percentage of live cells. N = 2 T cell donors with duplicate or triplicate technical replicates. Bars are averages ± SD.
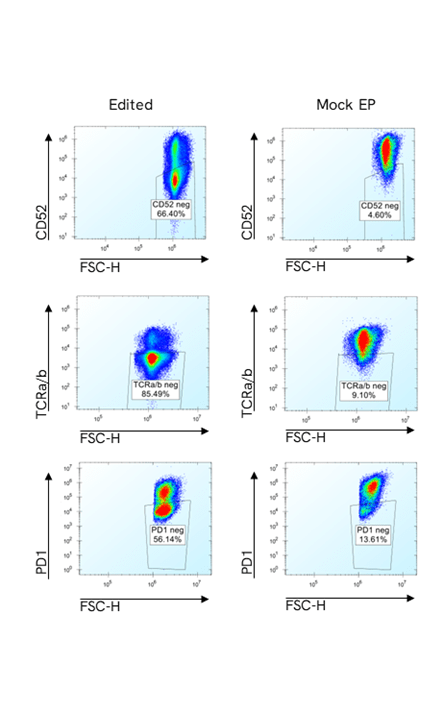
Figure 3
Representative flow cytometry plots from edited and control samples for each of the three markers. Negative cells for each marker are gated on the live population.
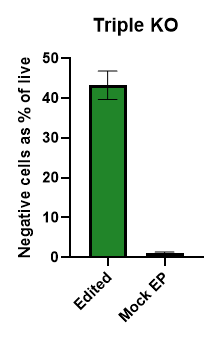
Figure 4
Percentage of individual cells that are negative for all three markers simultaneously (CD52, PD1 and TCRa/b) in the samples, edited with the Pin-point platform, and mock-electroporated (EP) controls, reported as percentage of live cells, with no additional enrichment. To induce the expression of PD1, T cells were cultured in the presence of phorbol 12- myristate 13-acetate and ionomycin for 48 h prior to analysis to enable simultaneous quantification of all three markers. N = 2 T cell donors with duplicate or triplicate technical replicates. Bars are averages ± SD.
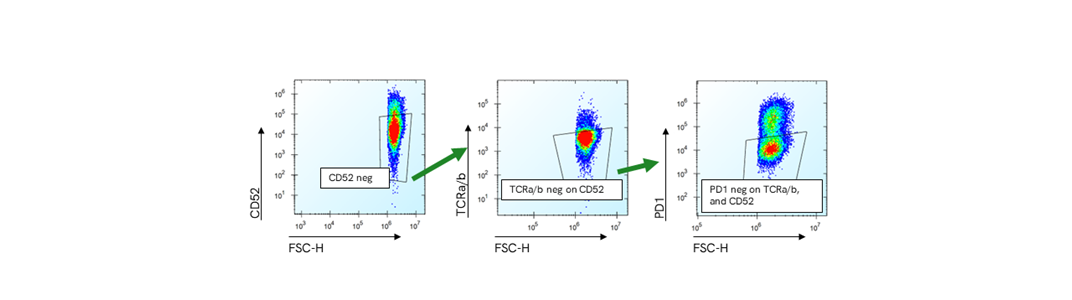
Figure 5
Representative flow cytometry plots from an edited sample to show the gating strategy: CD52 negative population gated on live cells; TCRa/b negative population gated on CD52 negative population; and PD1 negative population gated on CD52 and TCRa/b double negative population. The count of cells from this gate are the total number of cells negative for the three markers. The counts of triple negative cells are normalized on the counts of live cells and the percentage reported in the figure above.
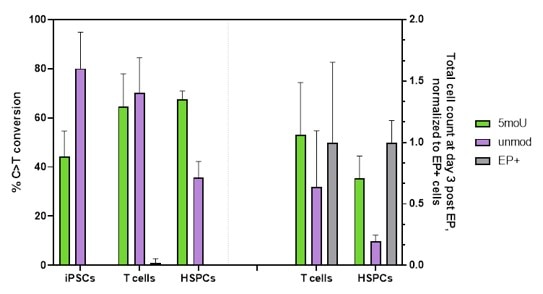
Figure 6
Representative editing and total cell count at 3 days post electroporation when delivering a Custom Pin-point Synthetic sgRNA targeting a chain of the major histocompatibility complex class 1 and either Pin-point unmodified or Pin-point 5moU modified mRNAs in T cells, iPSCs, and HSPCs.
Application notes
Posters
-
Performance and modularity of Revvity's Pin-point base editing system characterized by arrayed and pooled screening platforms - poster
-
Pin-point™ base editing system: a versatile editing platform driving cell therapies - poster
-
The Pin-point™ base editing platform streamlines the generation of hypoimmunogenic iPSCs for allogeneic cell therapy - poster
Quick protocols
Safety data sheets
Related Products
Non-targeting synthetic controls for evaluation of Pin-Point base editing. Bioinformatically designed to not target any gene in the human genome
Synthetic sgRNA controls to verify optimal parameters for precise gene editing without inducing DNA double strand breaks
The Pin-point base editing mRNA system allows for precisely directed point mutation edits without inducing double stranded breaks or the need for homology directed repair
Catalog ID:PNP12580
Unit Size:20 µg
