A revolution is under way in functional genomics which is spearheaded by the CRISPR-Cas9 system and its application to pooled genetic screening. Remarkable new tools, made possible by dCas9, are coming to fruition that will allow for a new kind of interrogation of gene function, allowing us to ask more sophisticated questions about the biology of drug targets.
January 2013 was marked by a major breakthrough in genome engineering. Four labs simultaneously engineered the bacterial and archaeal CRISPR-Cas9 system to induce precise cleavage at mammalian genomic loci1-4. Within a year's time, two back-to-back papers documented the application of CRISPR-Cas9 knockout technology to forward genetic screening5,6. These studies showed not just proof of concept of a new technology, but a spectacular jump in what is possible within functional genomics. Many studies have since capitalized on these discoveries and several publications, including from Horizon7, have demonstrated screening platforms with even greater precision and performance.
From gene editing to gene regulation
But knock-out is not the end! Inspired by modified versions of both ZnF nucleases and TALEs8, researchers have engineered a nuclease-dead version of Cas9 (dCas9). Rather than mutating specific genomic loci, dCas9 allows disruption of gene transcription simply by the binding of proximal sequences at the promoter region of the gene. This system, CRISPR interference (CRISPRi), efficiently blocks transcript initiation in E. coli and mammalian cells9-13. By covalently linking KRAB transcriptional repressor to dCas9, enhanced transcriptional silencing is achieved1,4.
Using the dCas9 approach, it is also possible to create a transcriptional activation tool, coined CRISPRa, by fusing dCas9 to the VP64 and p65 activation domains13,15-17. Effective gene activation with CRISPRa has been accomplished by several approaches: the SunTag array, which uses multiple VP64s recruited onto a peptide array18; VPR, a synergistic tripartite activation method using a fusion of VP64, p65 and Rta19; and the Synergistic Activation Mediator complex (SAM20), which uses a dCas9-VP64 and recruitment of p65 and HSF1 via RNA binding protein components. These adapted CRISPR tools provide incredible new opportunities to study gain-of-function mutations in genome-wide screens14,20.
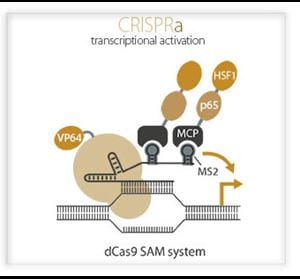
Horizon's CRISPRa Screening Tools
Power up!
For dCas9 to achieve full functionality, it must exert its activating or inhibitory effect on gene output through constitutive binding. Intriguingly, Hinz et al, Horlbeck et al, and Isaac et al discovered that dCas9 activity is influenced by nucleosome occupancy and that histone-DNA binding effectively blocks guideRNA access21-23. By coupling data available from multiple CRISPRi and CRISPRa screens to transcription start-site analysis (FANTOM), Horlbeck et al24 were able to generate highly optimised versions of their guideRNA design platforms. This advance in technology substantially increases the efficiency of these tools, and brings hit ID by pooled CRISPRi screening to a performance level which could exceed CRISPR knockout.
When is dCas9 the best?
With all these exciting new tools available, picking the right one for the right job is crucial for research success. In principle, knocking out genes rather than repressing their expression should provide the greatest potential window for discovery; however, the recent adaptations to CRISPRi guideRNA design appear to level the playing field here. CRISPR knockout isn't ideally suited to the study of hypomorphic phenotypes, including that of essential genes, and complete gene KO might not be the best model for druggability. But the high precision and penetrance of knockout technology provides unprecedented clarity and outstanding quality datasets.One chink in the armour of CRISPR knockout is the application to amplified loci, where multiple cuts can cause an off-target DNA damage response25,26. Also, since dCas9 does not alter the sequence of genomic DNA, its activity can be made inducible and reversible, which is not the case for the Cas9 nuclease. CRISPRi and CRISPRa approaches will also be more suitable for studying differential expression of long noncoding RNAs, genes that have proven difficult to target effectively with the CRISPR knockout platform. Finally and unique to CRISPRa is the exciting prospect to study gain-of-function phenotypes - this opens up myriad new paths to discovery.
With the greatly improved CRISPRi and CRISPRa technology enabling researchers to explore novel areas of biology normally out of reach with knockout technology, exciting times lie ahead.
Benedict Cross and Carlos Le Sage
- Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823-6 (2013).
- Jinek, M. et al. RNA-programmed genome editing in human cells. Elife 2013, (2013).
- Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-23 (2013).
- Cho, S. W., Kim, S., Kim, J. M. & Kim, J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230-232 (2013).
- Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic Screens in Human Cells Using the CRISPR/Cas9 System. Science (80-. ). 80, 1-8 (2013).
- Shalem, O. et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science (80-. ). 343, 84-87 (2014).
- Cross, B. C. S. et al. Increasing the performance of pooled CRISPR-Cas9 drop-out screening. Sci. Rep. 6, 31782 (2016).
- Beerli, R. R., Segal, D. J., Dreier, B. & Barbas, C. F. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. U. S. A. 95, 14628-33 (1998).
- Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173-1183 (2013).
- Larson, M. H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180-2196 (2013).
- Bikard, D. et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429-7437 (2013).
- Ji, W. et al. Specific Gene repression by CRISPRi system transferred through bacterial conjugation. ACS Synth. Biol. 3, 929-931 (2014).
- Gilbert, L. A. et al. XCRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, (2013).
- Gilbert, L. A. et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647-661 (2014).
- Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973-976 (2013).
- Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977-979 (2013).
- Farzadfard, F., Perli, S. D. & Lu, T. K. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2, 604-613 (2013).
- Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635-646 (2014).
- Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326-328 (2015).
- Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583-588 (2014).
- Hinz, J. M., Laughery, M. F. & Wyrick, J. J. Nucleosomes Inhibit Cas9 Endonuclease Activity in Vitro. Biochemistry 54, 7063-7066 (2015).
- Horlbeck, M. A. et al. Nucleosomes impede cas9 access to DNA in vivo and in vitro. Elife 5, (2016).
- Isaac, R. S. et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife 5, (2016).
- Horlbeck, M. A. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5, (2016).
- Aguirre, A. J. et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 6, 914-929 (2016).
- Munoz, D. M. et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 6, 900-913 (2016).
