After all the hard work of editing your cell line, you want to have confidence in your new research model. So, how do you verify your cell line is what you expect it to be? Could a heterogeneous cell population be obscuring your editing effects? Is observed phenotype being caused by the targeted gene edit, or unintended off-target effects?
Here we discuss ways to add supporting data to validate your gene-engineering projects.
Confirm the clonality
Follow these steps when you're ready to validate your clones:
1. Confirm the clonality of your cell by:
- Imaging
- Single cell PCRs
CRISPR gene edits leave a very heterogenous population, your gene editing event can give you a mixed population of cells with in-frame deletions, out-of-frame deletions, homozygous knockout populations and heterozygous knockout populations. If you test on this pool, you will have no idea what population is causing your desired effect. A good idea is to grow out clonal populations after your edit. Another top tip is to keep these different populations so that you have additional controls, or a second clone of the same genotype, so that you are confident in your result.
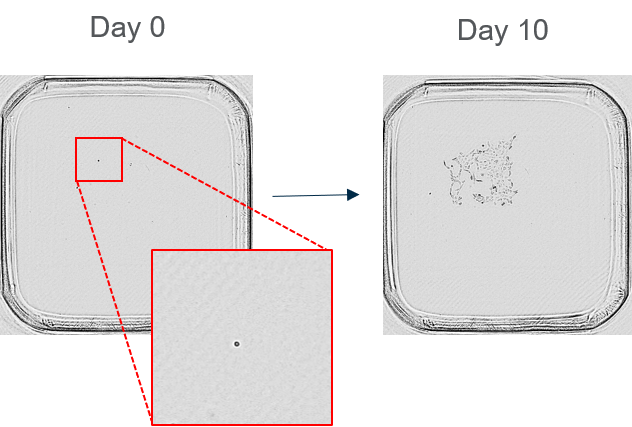
2. Confirm genotype with a suite of PCRs:
- Knockout
- Knock in
An important consideration to take when designing your PCRs is to design them so that you are looking for the genotype that you do not want. So, if you are looking for a homozygous knock-in or knockout genotype, really probe your cell population for residual WT allele. When you don’t find any wildtype, the sense of satisfaction is great.
3. Perform TOPO cloning to sequence individual alleles
A good way to really probe your genotype is to perform TOPO cloning. This is particularly important when you are dealing with those high copy number genes with 3 or more alleles. TOPO cloning makes it easy to separate the alleles from one another so you can fully understand the genotype of your cell.
4. Identify any off-target incorporation
The importance of identifying any possible off-targets will depend on how you are using the cells. Not all of us have the funding to perform next gen sequencing on every cell line we generate. One possible quick way of testing for off-targets is to sequence around the loci that were identified as having off-target scores in your guide RNA design. The next way to determine that your results are not due to an off-target effect is to use a second clone – which will hopefully display the same effect.
5. Confirm there is no contamination:
- Bacterial contamination
- Wildtype cell / non-targeted cell contamination
Now that you have confirmed the genotype, it is time to freeze a large stock of cells and make sure that they are not contaminated. A top tip is to freeze your cells, then revive a vial and check for bacterial contamination as well as WT allele or non-targeted allele contamination. It would be terrible to start working with this important cell line without realising they are contaminated. So, try to do this at this stage in your cell line generation to make sure that you are confident in all your future experiments.
6. Confirm and identify the suspected phenotypic changes to the cell; e.g some protein changes
You have a complete understanding of your genotype at this stage, but what does that mean to the protein that you have modified – how has it changed? Checking out the phenotypic changes too early in the cell line generation process can be a great idea, however do not be disheartened if you do not see your phenotypic change on the pool level. Quite often the phenotypic changes that you expect are masked at the pool level and are only visible after you have generated clonal populations.
Increase your confidence that the gene edits are the cause of the phenotype
Off-target effects are a common problem to be aware of. Ideally, your gene engineering work will have resulted in two clonal cell lines, produced by two separate guide RNAs, to increase confidence in your results.
The example below shows how you can use KO cell lines to interrogate your pathways of interest.
JAK-STAT signalling
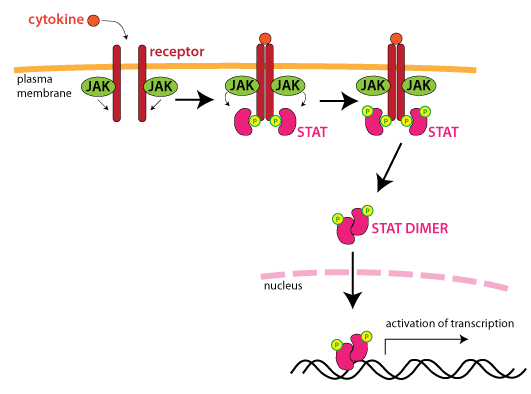
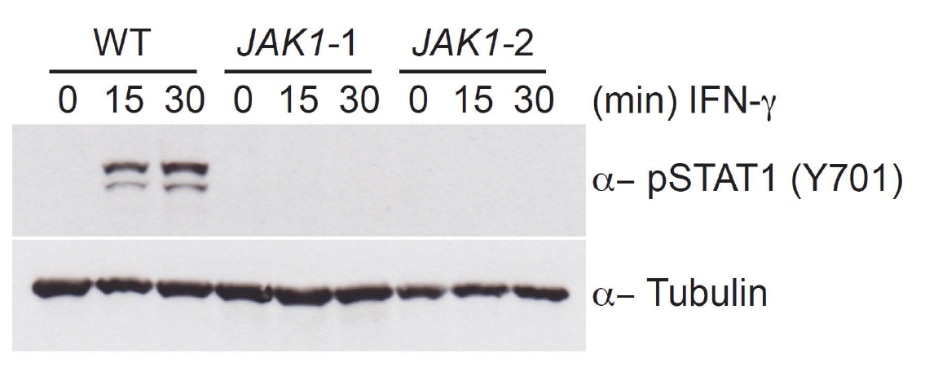
- Knockout of the gene of interest disrupts the cytokine response
- Phosphorylation of STAT1 is absent in JAK1 knockout clones
- Use of two clones, created by separate gRNAs, increases confidence that the effects are not due to off-target editing
Validation through gene rescue studies
The effect of possible off-target gene editing events has been a cause of much discussion and debate. To alleviate concerns, a phenotype can be rescued with simple molecular biology techniques.
This example involves the role of an E3 ligase called RFWD3. E3 ubiquitin ligases are known to have a role in DNA damage repair, particularly in homologous recombination (HR). The BRCA1 gene encodes an E3 ligase that is critical for HR and defects in the BRCA1 protein often lead to breast cancer.
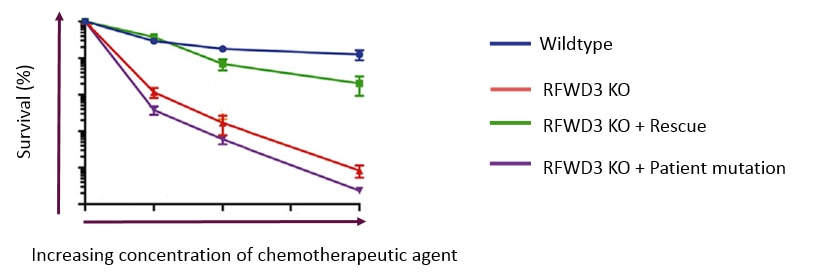
Here are the survival curves of HAP1 WT (blue) and the knockout (KO) RFWD3 (Red). The KO RFWD3 cells exhibited hypersensitivity to MMC (mitomycin C).
When the KO cell line is rescued by over-expression of the RFWD3 (transduced with a lentivirus expressing WT), the functionality is returned and the MMC sensitivity is reversed.
Interestingly, if instead of the wildtype, a protein with a patient relevant mutation is over-expressed, you can see the effects of these mutations on cell survival. In this case a patient relevant mutation with a particular point mutation, (a RING finger domain mutant, C315A) did not rescue the phenotype, demonstrating the connection between the genetic sequence and the disease mechanism .
Supporting experiments in a secondary model
Repeating key experiments in a separate cell line, with the same genetic perturbation, is another method to add independent support that your gene editing has been successful, as well as supporting your research hypothesis.
Horizon’s knockout Cell Lines
Horizon’s broad catalog of cell lines allows you to compare your gene of interest in an independent cell model, as well as up- or -downstream gene disruption in one highly characterised cell line background.
You can now access all our knockout cell lines from our Express Catalog, including:
- Collections of knockouts for probing entire pathways
- Number of clones available per knockout genotype
- Wildtype control included to validate your results
